Figures & data
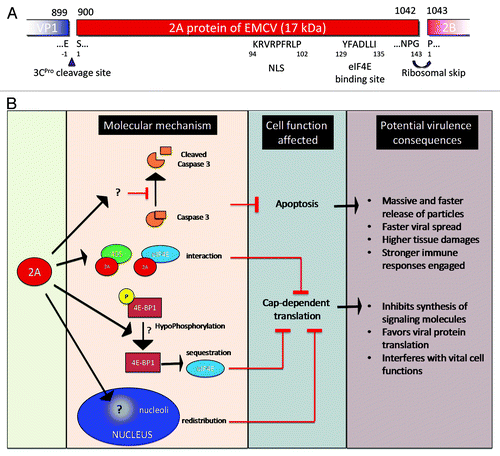
Figure 1. (A) Genomic organization. The EMCV genome is a positive single-strand RNA, covalently linked to the viral protein VPg in 5′. The genomic RNA is composed of two UTR in 5′ and 3′ and an ORF that encodes for a polyprotein (dark gray). The polyproteins is cleaved by the 3C protease, cleavage sites are indicated (black triangles). Ribosome skinping (curved right arrow). A second ORF (clear gray) due to a +2 ribosome frameshifting. cre, Cis replicating element; PKs, pseudoknots; IRES, internal ribosome entry site. (B) Description of EMCV proteins properties and functions known or assumed by similarity with other picornaviruses.
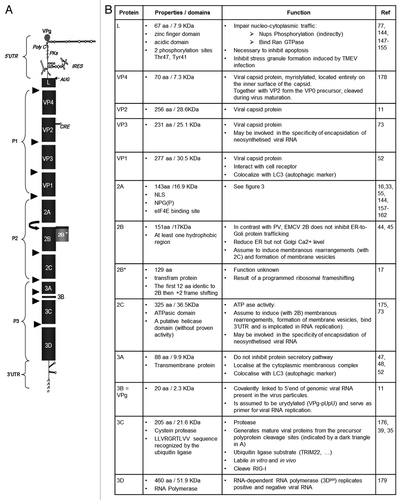
Figure 2. Encephalomyocarditis viral replication cycle. The virion binds to a cellular receptor. After uncoating, the genomic RNA is released by an unknown mechanism. Internalization (~1–2 h). Once in the cytoplasm, the VPg protein is detached to the 5′ end of the genome, the translation is initiated at the IRES that require the PTB cellular protein, the polyprotein is synthesized. Translation (~2.5–3 h). The polyprotein is cleaved during and after the translation, leading to precursors or mature proteins. Some of those proteins allow formation of the membranous vesicles where genome replication will occur. The positive genomic RNA is replicated into negative RNA thanks to the VPg-pUpU that serves as a primer for the 3D polymerase. The negative RNA syntesis leads to the production of a double stranded RNA molecule, the replication form (RF). The newly synthesized negative RNA in turn, serves as templates for synthesis of positive RNAs in the replication intermediate (RI). Replication (~3–4 h). Those new positive genomic RNAs will either serve for translation after removal of VPg, serve as matrix for synthesis of new viral RNAs or will be encapsidated. The viral capsid proteins VP0, VP1 and VP3 auto-assemble into a protomer. Five protomers will assemble into pentamers and 12 of them will form the icosahedric capsid. Encapsidation (~4–6 h). After RNA encapsidation, cleavage of the precursor VP0 into VP2 and VP4 allows maturation of the virion. Virions are then egress by cell lysis. Egress (~6–10 h). (Modified from ref. Citation177).
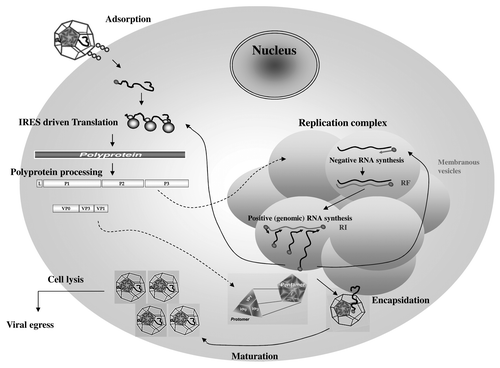
Figure 3. Encephalomyocarditis viral 2A protein. (A) EMCV 2A protein organization and putative sequence. (B) Functionality of EMCV 2A protein and potential consequence on EMCV virulence.