Figures & data
Figure 1. VP24 key features. (A) Purified EBOV1–233 VP24, SUDV1–233 VP24 and RESTV11–237 VP24 bind to purified STAT11–683 by ELISA. VP24s were coated onto the ELISA plate at 0.02 mg/ml for overnight incubation at room temperature. STAT11–683 was incubated on the plates, binding was detected with HRP-conjugated secondary antibody and the resulting O.D. was read at 450 nm. BSA was used as a negative control. (B and C) The overall shape of VP24 resembles a three-sided pyramid with Faces 1 and 3 as illustrated. A hydrophobic cavity in Face 1 and a polar cavity in Face 3 are indicated by arrows. A ribbon model of SUDV1–233 VP24 illustrates the underlying secondary structural elements, while a surface representation of SUDV1–233 VP24 indicates the sequence conservation. Navy indicates residues that are absolutely conserved among the ebola- and marburgviruses, while red indicates residues that are least conserved among these viruses. Face 2 is the least conserved of the three faces and is not illustrated. Sequence identity is calculated using Homolmapper.Citation37 The accession codes for SUDV1–233, SUDV11–233 and RESTV11–237 are 3VNE, 3VNF and 4D9O, respectively.
Figure 2. Possible STAT-1 interaction sites and RESTV-specific differences. (A) Results from deuterium exchange mass spectrometry analysis are mapped onto the structure of SUDV1–233 VP24. Peptidic segments colored blue exhibit slower H/D exchange, while segments colored red exhibit increased H/D exchange when in the presence of STAT11–683. Segments colored white exhibit no change. Segments colored gray were not observed in these experiments. Experimental details can be found in Zhang et al.Citation4 (B) Sequence conservation among only ebolaviruses (not marburgviruses) mapped onto SUDV1–233 VP24. Circles indicate the sites of change in H/D exchange upon binding to STAT11–683. (C–E) Sites where RESTV encodes amino acids different from the SUDV and EBOV are mapped in red onto the structure of RESTV11–237. It is unclear which of these differences are important. We note that many exist on the unconserved Face 2. Others map to sites already shown to be important for virulence. A cyan circle on Faces 2 and 3 outlines a region of VP24 shown to interact with karyopherin α1 (cyan) that is flanked by RESTV-specific residues R139 and S140. Black ovals on Face 3 outline regions of faster or slower H/D exchange in the presence of STAT1. Inside these regions are RESTV-specific residues T116, L107, S184 and T185. Residue S50, which has been shown to increase virulence of EBOV in mice,Citation36 is located on Face 1. Green arrows indicate the location of the Face 1 and 3 cavities.
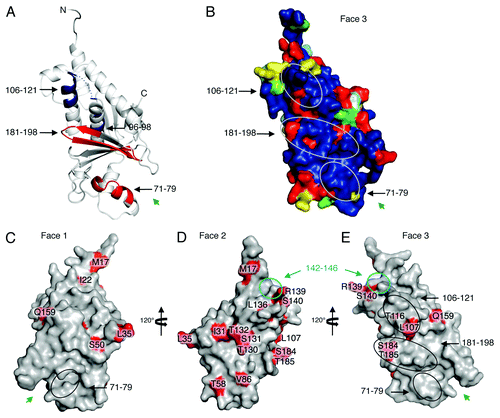
Figure 3. Excerpts from the ebolavirus operation plan. (A) Upon interferon activation, STAT1 becomes phosphorylated by JAK and forms either an ISG3 complex (STAT1-STAT2-IRF9) or a homodimer. The STAT1 complex is then translocated to the nucleus by the carrier protein karyopherin α1. VP24 targets karyopherin α1 to sequester STAT1 in the cytoplasm. VP24 may also be able to target STAT1 directly (interactions shown here in dashed lines), although the downstream result of this binding is not yet known. Inset shows that VP24 also interferes in the MKK3/6-p38 mitogen-activated protein (MAP) kinase pathway,Citation11 by blocking IFNβ stimulated phosphorylation of p38-α. (B) The presence of double-stranded RNA is detected by the sensors RIG-I and MDA-5 that then trigger signaling of mitochondrial antiviral signaling proteins (MAVS), also known as virus-induced signaling adapter (VISA), IPS-1 and Cardif to activate Tank binding kinase-1 (TBK-1) and I-Kappa-B kinase epsilon (IKKε). With the aid of TNF receptor-associated factor 3 (TRAF3), activated TBK-1 and IKKε then phosphorylate interferon regulatory factor (IRF) 3 and IRF7. Upon phosphorylation, IRF3 and IRF7 homodimerize, translocate to the nucleus, and subsequently initiate production of type I IFNs. VP35 binds and masks dsRNA to prevent this signaling and downstream expression of IFNs. In addition, VP35 can also inhibit the phosphorylation of IRF3Citation21 by interacting with the kinase domain of IKKε22. In this way, both VP35 and VP24 may interfere at multiple steps in their respective pathways.