Figures & data
Figure 1. Molecular phylogenetic tree generated in this study by Maximum Likelihood based on Kimura 2- parameter (K2P + G) for CAL data set (A) and Tamura 3-parameter (T92 + I) for ITS1/2 + 5.8S data set (B) of S. schenckii complex species. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (Bootstrap support values > 70% are indicated in bold). GenBank accession numbers are indicated next to strain code. Strains used in this study are indicated in bold type.
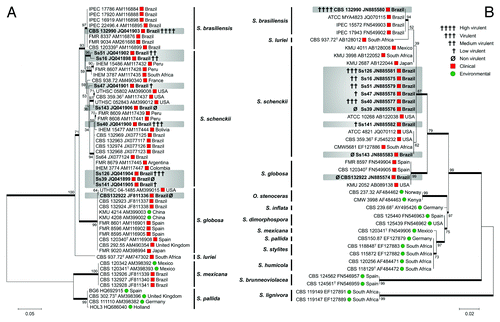
Figure 2. Survival of mice infected with Sporothrix spp. p < 0.05 for Ss54 (CBS 132990) and Ss40 vs. other isolates; p < 0.05 for Ss16, Ss51, Ss126 and Ss141 vs. other isolates and p < 0.05 for Ss143, Ss47, Ss39 and Ss06 (CBS 132922) vs. other isolates.
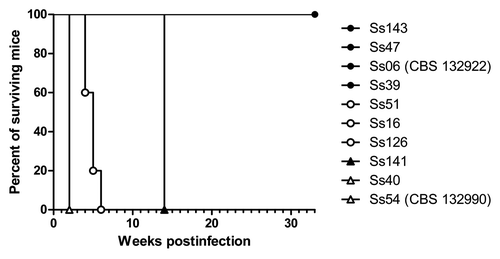
Figure 3. Colony count (CFU/g) comparison among different strains of S. schenckii s. str., and S. brasiliensis or S. globosa in spleen (A), lungs (B) and liver (C) obtained from infected BALB/c mice. Wide bars represent the mean CFU/g of tissue recovered from five mice and thin bars represent the standard deviation. *p < 0.05.
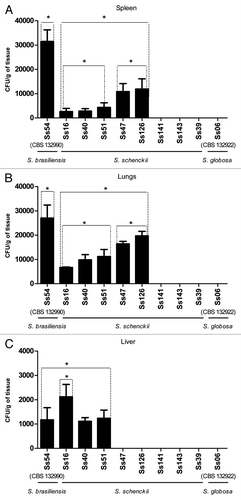
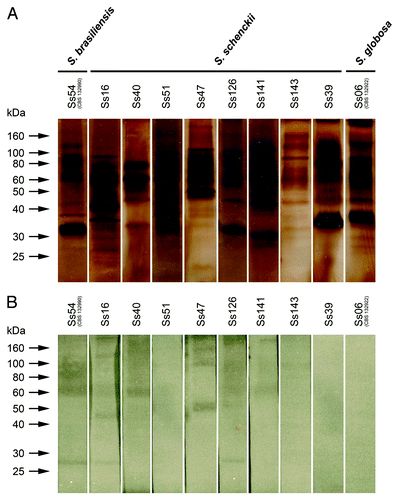
Figure 4. (A) Protein/glycoprotein profile of exoantigens of S. schenckii, S. brasiliensis and S. globosa, detected by SDS-PAGE. (B) Immunogenic molecules from exoantigens recognized by antisera from infected animals—western blot assay. S. brasiliensis isolate Ss54 (CBS 132990); S. globosa isolate Ss06 (CBS 132922) and S. schenckii s. str. isolates Ss16, Ss39, Ss40, Ss47, Ss51, Ss 126, Ss141 and Ss143.