Figures & data
Figure 1. Images of AGE-modified Salmonella bearing different bacterial surface appendages. (A) Depicted is an image of S. Typhimurium P1-pHCCitation34 overexpressing ETEC CFA/I fimbriae as observed by atomic force microscopy. Narrow blue arrows indicate CFA/I fimbriae while bold white arrows indicate flagella. (B) Depicted is an image of S. Typhimurium P1-pHFCitation39 overexpressing Yersinia pestis F1 capsule as observed by field emission microscopy. The capsule is labeled with 20 nm immunogold particles (black spots). Narrow yellow arrows indicate the capsular proteins secreted by the cell forming a mushroom-like structure, which is in the early stage of capsule formation. Bold black arrows indicate the bacterial cell. (C) Depicted is an image of S. Typhimurium P1-pTP2DC+CCitation40 expressing flagella as observed by field emission microscopy. Narrow white arrows indicate flagella while bold yellow arrow indicates bacterial cell. (D) Depicted is an image of wt S. Typhimurium as observed by atomic force microscopy.
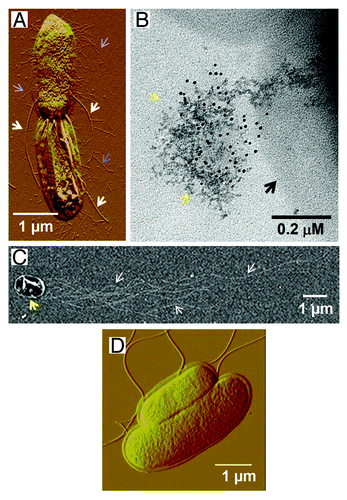
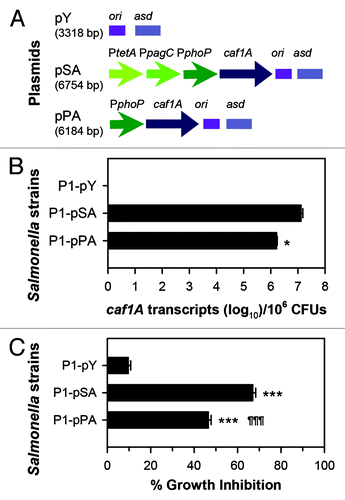
Figure 2. The extent of usher gene caf1A expression correlates to the amount of channel formation. (A) Schematic physical maps of asd-based plasmids are depicted: the caf1A gene is regulated by a tripartite fusion promoter in P1-pSACitation39 or a single promoter in P1-pPA; and the vector control lacking caf1A is harbored in P1-pY. (B) The expression of caf1A was determined by real-time polymerase chain reaction (RT-PCR). pSA DNA was used as a standard. The bacterial strains P1-pSA, -pPA, and control -pY were grown in LB medium at 37 °C with shaking at 200 rpm. Cells in log phase were harvested for RNA purification and verification of the number of transcripts using protocols according to the manufacturers' instructions (Invitrogen). The Student t-test was used to determine the difference of caf1A mRNA copy numbers between P1-pSA and -pPA, with *P < 0.05. (C) The Caf1A channel formation was determined by bacterial growth inhibition in the presence of erythromycin. The three strains P1-pSA, -pPA, and -pY were inoculated to LB medium at an optical density at 600 nm (OD600) of 0.2 with or without the supplementation of 64 µg/ml erythromycin. At three hours post-inoculation, cell OD600 was measured. Growth inhibition was calculated by using a formulation: 100 × (1 - OD600 with erythromycin/OD600 without erythromycin)%. The Tukey multiple comparison test was used to determine difference of growth inhibition among the three strains; ***P < 0.001 indicates the growth inhibition of P1-pSA or -pPA vs. -pY, and ¶¶¶P < 0.001 indicates the growth inhibition of -pPA vs. -pSA. Experiments for both (B) and (C) were performed three times. Presented data are the means ± SD.