Figures & data
Figure 1. Alcohol (EtOH) inhibits J774.16 macrophage-like cells binding and phagocytosis of Acinetobacter baumannii (Ab). J774.16 cells were untreated or exposed to EtOH for 2 h followed by incubation with Ab. (A) Binding and phagocytosis of FITC-labeled Ab by J774.16 cells was determined using fluorescent activated cell sorting (FACS) analysis. Representative plots of bound and internalized FITC-labeled bacteria by macrophage-like cells are shown. Each plot was generated after 10 000 events were analyzed. (B) Light (LM) and fluorescent microscopy images of J774.16 cells phagocytosis of FITC (green fluorescence)-labeled Ab after exposure to 12.5 mmol/L EtOH. Scale bar: 10 μm. The experiments were performed twice with similar results obtained.
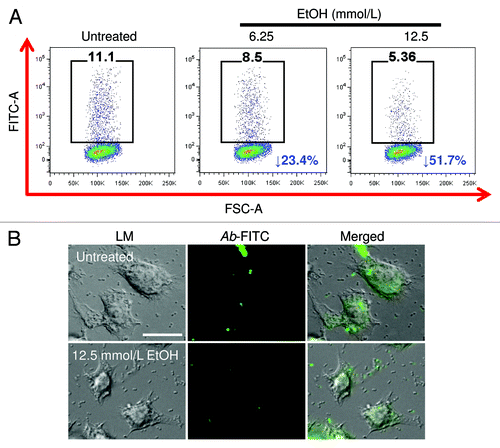
Figure 2. EtOH impairs complement receptor 3-mediated phagocytosis. (A) Western blot (WB) analyses of CD11b, CD18, and RhoA of untreated and EtOH (6.25 or 12.5 mmol/L)-treated J774.16 cells. (B) Quantitative measurements of individual band intensity in WB in (A) for CD11b, CD18, and RhoA using Image J software. GAPDH was used as a loading control to determine the relative intensity ratio. Bars are the averages of three independent gel results, and error bars denote standard deviations. Asterisks denote a reduction (*P < 0.001) in the intensity of the band of RhoA compared with GAPDH. The experiments were performed thrice with similar results obtained.
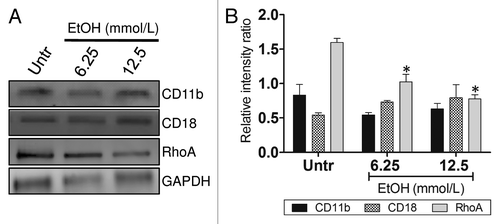
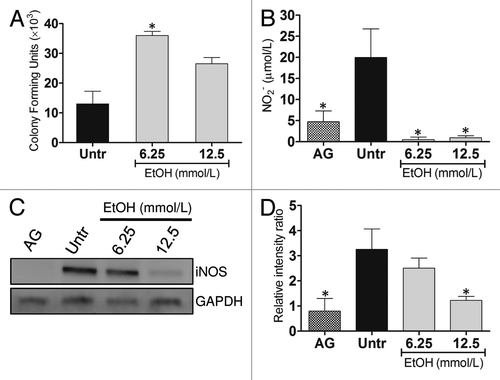
Figure 3. EtOH reduces Ab killing by J774.16 cells. (A) Killing of Ab by J774.16 cells was determined using colony-forming units (CFU) analysis. (B) EtOH inhibits nitric oxide (NO) production by macrophage-like cells. NO production was quantified using the Griess method after J774.16 were untreated or EtOH-treated and co-incubated with LPS for 96 h. For (A and B), bars and error bars denote average of three measurements and standard deviations, respectively. Asterisks denote P value significance (*P < 0.001) calculated by analysis of variance and adjusted by use of the Bonferroni correction. (C) WB analysis of inducible nitric oxide synthase (iNOS) of untreated, aminoguanidine (AG; iNOS inhibitor)-, and EtOH-treated J774.16 cells. (D) Quantitative measurements of individual band intensity in WB in (C) for iNOS. GAPDH was used as a control to determine the relative intensity ratio. Bars are the averages of three independent gel results, and error bars denote standard deviations. Asterisks denote a reduction (*P < 0.001) in the intensity of the band of iNOS compared with GAPDH. The experiments were performed thrice with similar results obtained.