Figures & data
Figure 1. Unfolded protein response (UPR) pathways in eukaryotes. (A) The mammalian UPR pathway consists of three ER-transmembrane sensor proteins, IRE1, PERK, and ATF6. Activation of IRE1 cleaves the 26 nt intron of XBP1u mRNA, and the activated XBP1s bZIP transcription factor upregulates many essential UPR target genes. On the other hand, the translated XBP1u protein appears to sequester XBP1s protein in the cytosol. IRE1 also controls selective mRNA decay (RIDD). Activation of PERK blocks general protein synthesis and increases the specific translation of ATF4 mRNA via phosphorylation of eIF2α. The ATF4 bZIP transcription factor induces expression of UPR target genes. ATF6 is a type II ER transmembrane protein with a bZIP domain. Upon ER stress, ATF6 is translocated to the Golgi and processed proteolytically by site-1 protease (S1P) and site-2 protease (S2P); the ATF6 fragment with the bZIP domain (ATF6f) is then released and translocates to the nucleus to activate UPR genes. (B) The UPR pathway in Arabidopsis thaliana consists of two branches: one involving endoribonuclease IRE1 and the other involving the proteolytic processing of membrane-associated bZIP transcription factors (bZIP17/28). Upon ER stress, IRE1 removes the 23 nt intron of bZIP60 mRNA, resulting in a bZIP protein lacking a transmembrane domain (bZIP60s) via frameshift translation. The bZIP60s transcription factor translocates to the nucleus to activate UPR target genes. Similar to mammalian ATF6, the membrane-associated bZIP transcription factors (bZIP17/28) are processed at the Golgi by S1P and S2P, releasing the truncated versions of bZIP17/28 into the nucleus to activate UPR target genes. Regulated IRE1-dependent decay of specific mRNAs in Arabidopsis has also been observed recently. (C) The yeast Saccharomyces UPR pathway is composed of the Ire1 kinase and the Hac1 bZIP transcription factor. Accumulation of unfolded or misfolded proteins in the ER lumen causes Ire1 to dimerize and trans-autophosphorylate through its kinase domain. The activated Ire1 kinase removes the unconventional intron (252 nt) of the HAC1 mRNA and a tRNA ligase, Rlg1, joins the two exons without the help of conventional spliceosomes. Spliced HAC1 mRNA is translated to produce an active Hac1 protein, which translocates to the nucleus to upregulate expression of UPR target genes encoding ER-resident chaperones and other proteins. “K” and “R” in Ire1 indicate the kinase and ribonuclease domains, respectively.
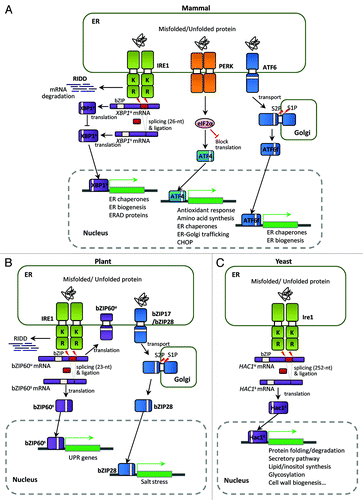
Figure 2. Conservation of the putative unconventional splicing sites of HXL1 homologs in basidiomycetes. (A) The unconventional intron sequences of HAC1/XBP1/HXL1 homologous mRNAs are aligned. Flanking exon sequences are denoted by uppercase letters and intron sequences by lowercase letters. Length indicates nucleotide length of the non-conventional intron that can be removed by Ire1. The conserved sequences of 5′- and 3′- splicing junctions are represented by sequence logo and dotted box. (B) Predicted secondary mRNA structures of HXL1 homologs in some basidiomycetous fungi. Putative splicing sites in 5′ and 3′ intron borders are located in the loop regions of stem-loop structures. The putative Ire1-mediated splicing sites and introns are indicated with arrowheads and written in lower case, respectively. Conserved sequences in splicing junctions are indicated by dotted boxes. Alignment and RNA secondary structure prediction were performed with the CLC main benchwork 6.8.4 (CLC bio). Nucleotide sequences were retrieved from the NCBI database and fungal genomics resource at JGICitation61: Cryptococcus neoformans (HXL1, CNAG_06134), Trichosporon asahii (A1Q2_03745), Tremella mesenterica (TREMEDRAFT_57223), Tremella fuciformis (bZIP1, GU723640.1), Dioszegia cryoxerica (fgenesh1_kg.80_#_88_#_Locus1962v1rpkm301.65), Yarrowia lipolytica (HAC1, XM_500811.1), Aspergillus nidulans (hacA, AJ413273), Aspergillus fumigatus (hacA, XM_743634), Trichoderma reesei (hac1, AJ413272), Alternaria brassicicola (HacA, AB01954.1), Candida albicans (HAC1, EF655649), Pichia pastoris (HAC1, FN392319), Saccharomyces cerevisiae (HAC1, NC_001138.5), Homo sapiens (XBP1, NM_005080), Caenorhabditis elegans (xbp-1, AF443190), Arabidopsis thaliana (bZIP60, AY045964).
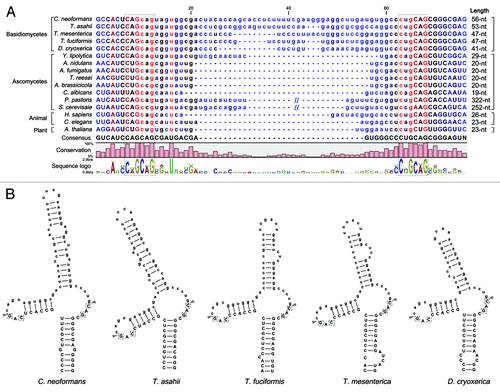
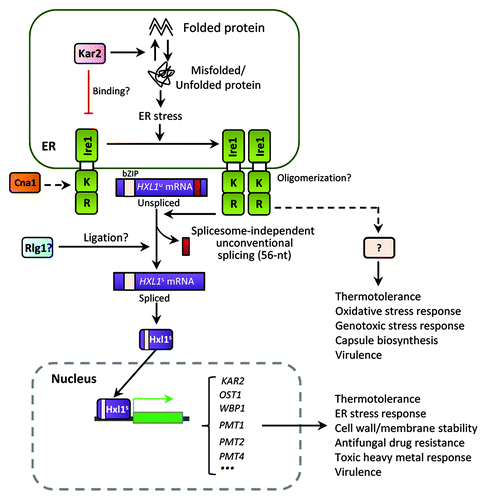
Figure 3. The ER stress response and UPR pathways in C. neoformans. The Cryptococcus UPR pathway consists of the Ire1 kinase, a bZIP transcription factor Hxl1, and their target genes. Upon ER stress, the spliceosome-independent unconventional splicing event in HXL1 mRNA occurs. Activated Hxl1 translocates to the nucleus and induces the expression of UPR target genes such as KAR2, which encodes an ER-resident molecular chaperone. The UPR pathway plays Ire1/Hxl1-dependent roles in ER stress response, antifungal drug resistance, and virulence. However, Ire1 also appears to have Hxl1-independent functions. Crosstalk between the UPR and calcineurin pathways via Cna1 is also indicated in Cryptococcus. Black arrows represent positive regulation or activation whereas red barred lines indicate negative regulation or repression. Dotted arrows indicate potential or unclear regulation.