Figures & data
Figure 1. An overview of the main methods of novel gene discovery using metagenomics. A sample from the environment of interest (e.g., soil, human gastrointestinal tract, or ocean) is collected (A), total metagenomic DNA is isolated directly or indirectly from the sample either by harsh or gentle lysis (B); metagenomic DNA is subsequently subjected to sequence-based analysis (C), which can include creating a clone library followed by random Sanger sequencing or direct sequencing of metagenomic DNA using next generation (NGS) methods (C-1), gene targeting using degenerate primers designed from conserved regions of homologous sequences followed by PCR amplification (C-2), data mining of existing metagenomic sequence data sets for genes or conserved domains of interest, followed by complete synthesis of host-optimized genes (C-3); capture of novel genes from plasmids using the TRACA (transposon-aided capture) method or integron-associated genes using PCR with primers that target conserved integration site sequences (C-4). Functional metagenomics (D) involves the creation of a small- or large-insert library using a specific type of vector (plasmid, fosmid, cosmid, or bacterial artificial chromosome [BAC]) depending on the needs of the user or aims of the project (D-1), choosing a suitable heterologous host for expression of metagenomic DNA. The most widely used host is E. coli, but species of Streptomyces and Bacillus, for example, have also been utilized. The library is maintained in 96- or 384-well micro-titer plates and stored at –80 °C (D-2). Clones containing metagenomic DNA can be screened to identify a phenotype of interest by replicating all or a portion of the library into new micro-titer plates or onto agar plates. Growth of library clones can be assessed for production of antimicrobials, pigmentation, altered morphology, or increased resistance to various stressors relative to control strains carrying empty vectors (D-3). Transposon mutagenesis may be performed on positive clones to identify the gene or genes of interest (D-4), which may be subsequently cloned as an isolated gene(s) to confirm the phenotype (D-5). Novel genes are discovered using sequence-based or functional metagenomic strategies, or a combination of both (E).
![Figure 1. An overview of the main methods of novel gene discovery using metagenomics. A sample from the environment of interest (e.g., soil, human gastrointestinal tract, or ocean) is collected (A), total metagenomic DNA is isolated directly or indirectly from the sample either by harsh or gentle lysis (B); metagenomic DNA is subsequently subjected to sequence-based analysis (C), which can include creating a clone library followed by random Sanger sequencing or direct sequencing of metagenomic DNA using next generation (NGS) methods (C-1), gene targeting using degenerate primers designed from conserved regions of homologous sequences followed by PCR amplification (C-2), data mining of existing metagenomic sequence data sets for genes or conserved domains of interest, followed by complete synthesis of host-optimized genes (C-3); capture of novel genes from plasmids using the TRACA (transposon-aided capture) method or integron-associated genes using PCR with primers that target conserved integration site sequences (C-4). Functional metagenomics (D) involves the creation of a small- or large-insert library using a specific type of vector (plasmid, fosmid, cosmid, or bacterial artificial chromosome [BAC]) depending on the needs of the user or aims of the project (D-1), choosing a suitable heterologous host for expression of metagenomic DNA. The most widely used host is E. coli, but species of Streptomyces and Bacillus, for example, have also been utilized. The library is maintained in 96- or 384-well micro-titer plates and stored at –80 °C (D-2). Clones containing metagenomic DNA can be screened to identify a phenotype of interest by replicating all or a portion of the library into new micro-titer plates or onto agar plates. Growth of library clones can be assessed for production of antimicrobials, pigmentation, altered morphology, or increased resistance to various stressors relative to control strains carrying empty vectors (D-3). Transposon mutagenesis may be performed on positive clones to identify the gene or genes of interest (D-4), which may be subsequently cloned as an isolated gene(s) to confirm the phenotype (D-5). Novel genes are discovered using sequence-based or functional metagenomic strategies, or a combination of both (E).](/cms/asset/a8f91555-4619-4b3a-b9ac-37f11fb870cf/kvir_a_10927208_f0001.gif)
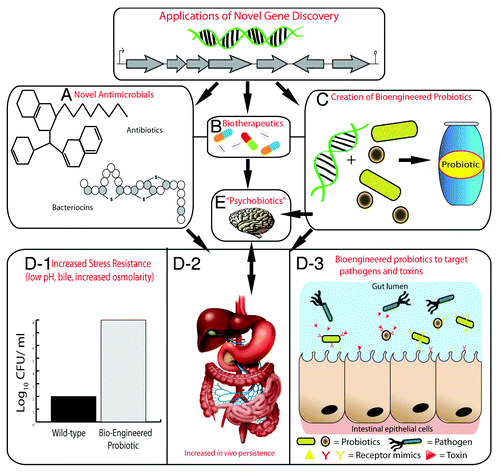
Figure 2. Applications of novel genes discovered by metagenomics. Novel genes or gene clusters discovered through metagenomics will encode proteins for the production of novel antimicrobial compounds such as antibiotics and bacteriocins (A) and biotherapeutics (B). Novel genes may also be used to create bioengineered probiotic strains (C). Bioengineered probiotics can be created that are more resistant host-associated stresses of the gastrointestinal tract such as low pH, bile, and increased osmolarity (D-1), which will lead to increased survival and persistence in the gastrointestinal tract (D-2). This will ultimately increase their therapeutic efficacy; using them to target specific pathogens or toxins by expressing receptor-mimic structures on their surface, thus preventing infection (D-3). The use of probiotics that have a health benefit to persons with a psychiatric illness have been termed “psychobiotics”. The creation of bioengineered psychobiotics that can produce neuroactive compounds at an increased level or for a specific illness may be possible by identifying novel genes through metagenomics (E).