Figures & data
Figure 1. Detection and chromatographic characterization of biofilm-degrading activity from L. gummosus. (A) Crude extracts prepared from the extracellular material of different Lysobacter species grown for 3 d on solid medium were tested against S. epidermidis biofilms on 24-well plates. Lant, L. antibioticus; Lenz, L. enzymogenes (DSMZ 2043); Lgum, L. gummosus; Lsp, Lysobacter sp. (DSMZ 3655); Lbru, L. brunescens; C water control. (B) L. gummosus extract was loaded onto a Q Sepharose Fast Flow column, and eluted isocratically with 100 mM NaCl (fractions 1–5). The flow-through (FT), the wash fractions (W), and the elution fractions were tested for biofilm-degrading activity. (C) The active fraction from the Q Sepharose column was passed over a Mono S 5/50 GL column and subsequently loaded on a Mono Q 5/50 GL column. Fractions 1–18 were eluted with 0–225 mM NaCl and tested for biofilm-degrading activity.
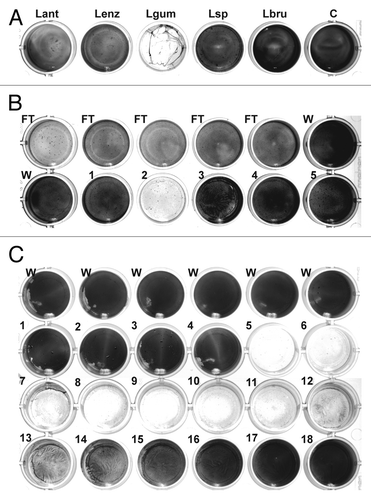
Figure 2. Purification of biofilm-degrading activity for peptide mass fingerprinting. (A) The fractions obtained by chromatography of the L. gummosus extract on a ProPac-SAX10 column were tested for biofilm-degrading activity (ST, starting material; FT, flow-through; W, wash; C, water control). (B) The fractions were analyzed by SDS-PAGE followed by staining with Flamingo Fluorescent Gel Stain. The bands labeled 1, 2, and 3 were excised and digested in-gel with trypsin for peptide mass fingerprinting. Fractions 8 and 17 were digested in solution with trypsin for peptide mass fingerprinting.
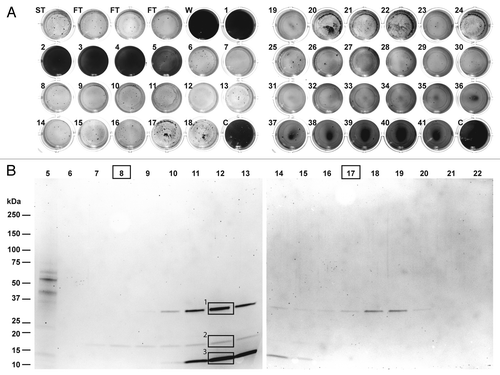
Table 1.L. gummosus proteins that co-purified with biofilm-degrading activity
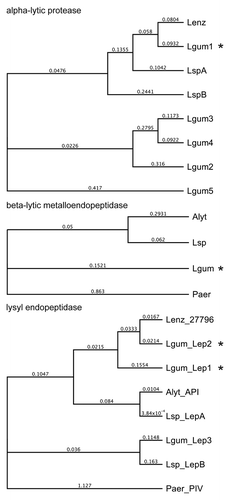
Figure 3. Phylogeny of L. gummosus enzymes that co-purified with biofilm-degrading activity. The phylogenetic trees were constructed on the basis of the amino acid sequences of the predicted preproenzymes using Geneious Tree Builder software (Blosum62; gap open penalty, 3; gap extension penalty 3). Alyt, Achromobacter lyticus; Lenz, L. enzymogenes; Lgum, L. gummosus; Lsp, Lysobacter sp.; Paer, Pseudomonas aeruginosa. The proteins in the fractions with biofilm-degrading activity are marked with an asterisk. Accession numbers are listed in the supplemental material.