Figures & data
Figure 1. The genomic organization of different types of LTR-retrotransposon families present in C. albicans. RT, reverse transcriptase DNA polymerase domain; RH, reverse transcriptase RNase H domain; PR, proteinase; IN, integrase.
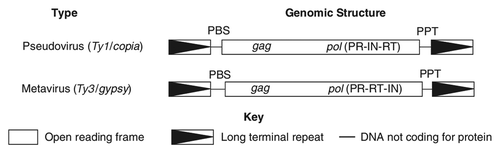
Table 1. Properties of C. albicans retrotransposon LTR families
Figure 2. LTRs can be subdivided into three regions: U3, R, and U5. U3 contains the enhancer and promoter sequences that drive viral transcription. R domain encodes the 5′ capping sequences (5′ cap) and the polyA (pA) signal.
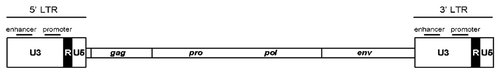
Figure 3. The diversity of PBSs in C. albicans retrotransposons. The names of the retrotransposons and the associated LTRs are shown on the left. LTR sequences are underlined. The GenBank accession numbers of the tRNAs are as follows: tRNAArg, AF041470; tRNAiMet, AF069449; tRNAIle, Y08492; tRNAGln, AF180282; and tRNAAla, Y08493.
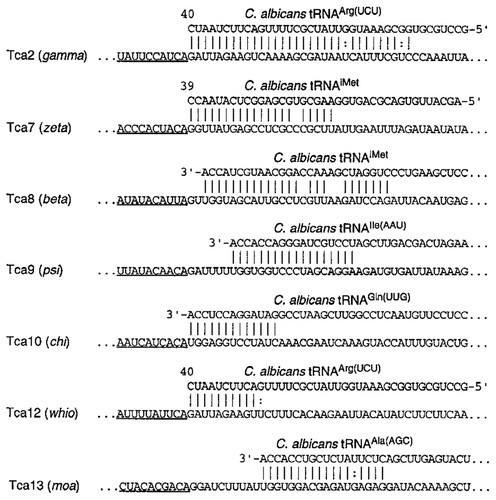
Figure 4. Mechanisms of retrotranspositions. The RNA transposition intermediate (brown) of an LTR retrotransposon provide plus strand RNA. The RNA has a tRNA (blue) base-paired sequence, the PBS, near its 5′ end (1). Primer tRNA anneals to binding site on RNA (2). The first sequences to be copied are the unique sequence at the 5′ end of the RNA (U5, red) and a short repeat sequence (R, green) present at both ends of the RNA. In this step, single-stranded DNA R region pairs with 3′ terminus in the first jump (3). Reverse transcriptase starts synthesis minus strand DNA (4), starting with U3 (purple) adjacent to R, and continuing until U5 and R are copied a second time. tRNA primer is removed. The RNA template is degraded by RNase H leaving a fragment at the poly-purine tract to prime second strand DNA synthesis (5). In the second jump, reverse transcriptase transfers to the other end of minus strand (7). Synthesis then proceeds (8). Integrase inserts this into chromosomal DNA, and transcription initiating in one LTR and terminating in the other generates genomic RNA with terminal repeats (9).
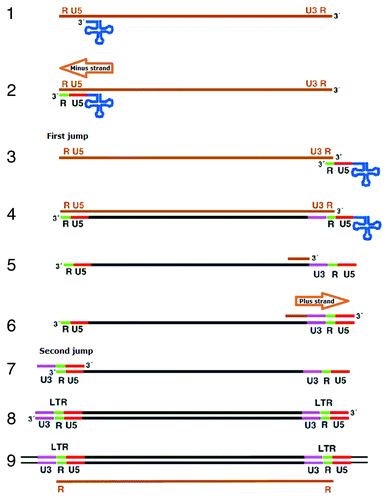
Figure 5. Southern blot analysis of DNA from lambda clones CJY-3 and CJY-4. Genomic DNA from strain SC5314 or purified DNA from lambda clones CJY-3 and CJY-4 was digested with EcoRI and hybridized with the α-element probe.
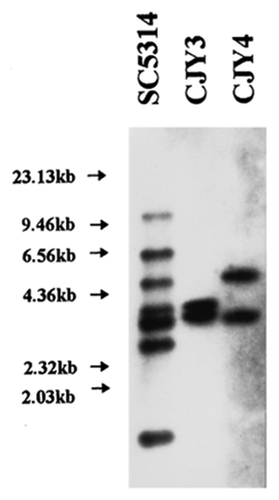
Figure 6. Restriction site maps of the genomic inserts from lambda clones CJY-3 and CJY-4 containing Tca1–1 and Tca1–2, respectively. The locations of elements (LTRs) are indicated by the boxed regions. Also indicated are the plus- and minus-strand primer binding sites (1PBS and 2PBS), the 5 bp direct repeats flanking the elements, the EcoRI fragments associated with each locus, and the 1.8 kb HindIII-EcoRI and 2.2kb EcoRV fragments that are used together as a hybridization probe of the internal region. The direction of transcription of Tca1 is from left to right.
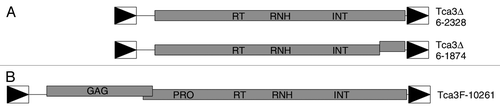
Figure 7. Structures of Tca3 elements. (A) The two Tca3 elements in Assembly 6 of the Stanford C. albicans sequencing project database. (B) A full-length Tca3 element from C. albicans strain ATCC10261. In all panels shaded boxes represent the ORFs of the elements. The locations of the conserved domains are indicated. Offset boxes and vertical lines within the boxes represent frameshifts and premature stop codons, respectively. The LTRs are represented by the boxed triangles. The common scale is shown at the bottom.