?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Introduction: EU Regulations continue to be strict in order to allow the highest degree of insurance in foodstuffs’ safety and quality. The traceability and labeling of imported products in EU still remain a valuable issue (EU-Regulation 178/2002). One of the most important concerns of the customers is the product traceability, which is defined as the ability to screen the whole history or geographical origin or farming type of any food by certified methodologies. In this regard, we proposed to study the link between microbial ecology and farming type of foodstuffs via a unique molecular analytical technique which is followed by an image analysis.
Purpose: A unique molecular analytical tool using 16S rDNA and 26S rDNA profiles generated by “PCR-DGGE” was used to detect the variation in bacterial and yeast communities respectively of peaches from Jordan (control fruit, organic farming and conventional farming).
Results: When the 16S and 26S rDNA profiles were analyzed using image and multivariate analysis, certain microbiota were detected on peach fruits originating from Jordan. The resulted band profiles of bacteria and yeasts from different farming types were specific and could be used as a bar-code to discriminate the fruits’ farming type.
Significance of the paper: This method could be considered as a new traceability technique which provides fruits with a unique biological bar-code and permits the possibility to control the farming type of foodstuffs.
1 Introduction
Food safety is considered nowadays as a critical issue for the imported food to EU. EU regulation imposed the traceability to all imported kinds of food. Over a long period of time, food industry had very simple traceability systems, but after increasing implementation of current Good Manufacturing Practice (GMP), the traceability systems have become important in the production chain. Traceability can be defined as the ability to retrieve the history or geographical origin of an article or its relevant components or an activity through a registered method for standardization (ISO, 2007). Installing these documentary systems in developing countries like sub-Saharan Africa is facing a lot of difficulties that is why new traceability strategies have been developed. In order to follow the farming types of peach fruit products during processing, we proposed to identify and validate some linked and important biological markers that come from the original habitat of these fruits.
The peach (Prunus persica) is defined as a deciduous tree that is native to Northwest China, in the region that present between the north slopes and Tarim Basin of the mountains of Kunlun Shan, where it was first cultivated [Citation1]. It has an edible juicy fruit that is called a peach. The name of the species P. persica is linked to its spread cultivation in Persia. It also belongs to the genus Prunus that includes the cherry and plum in the family Rosaceae. Finally, the peach is classified with the almond in the subgenus Amygdalus that is distinguished from the other subgenera by the corrugated shell of seed.
The most common analytical methods, which allow ensuring the determination of farming type, used bar-code, stable isotope, spectroscopy, etc. [Citation2]. It seems difficult to use the genomic markers of the fruit to ensure the traceability of peaches. However, skin of fresh fruits is contaminated and not sterile, so it may carry microorganisms or any of their DNA fragments. The occurrence of various microorganisms depends on the fruits' external environment (spoilage, insects, soil ecology, diseases etc.), but also microorganisms introduced by human activities [Citation3].
The idea was to develop a “biological bar code” that is based upon analysis of the DNA of micro flora present on the products [Citation4]. This technique is based upon the postulation that fruits' microbial communities are specific for a certain farming type.
The main motive of this paper was to perform PCR-DGGE, a method to analyze in a specific unique step all the bacteria and yeasts that present on the fruit in order to create an analytical technique that allows the linkage of bacterial and yeast communities to the farming type and avoid any individual analysis of each microbial strain. To the best of our knowledge, “this paper describing a molecular method of microbial ecology”, the PCR-DGGE that allows the certification of peach fruits using 16S rDNA and 26S rDNA fingerprinting of bacteria and yeasts, is one of the first techniques that link microbial ecology to fruit processing. Only Hamdouche et al. [Citation5] described a similar method on cocoa during fermentation.
The most specific advantage of this technique is that it allows analysis for both non-cultivable and cultivable, anaerobic and aerobic bacteria and also provides a rapid analytical methodology to observe any changes in the community structure in response to different environmental factors [Citation6].
2 Materials and methods
2.1 Sampling of fruits
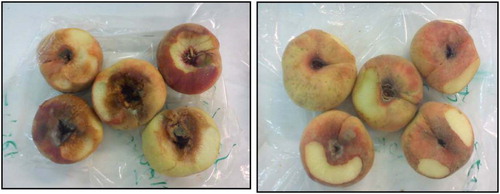
Mature peach fruits were harvested and collected from two different farming types in Jordan, which are organic and treated fruits from conventional farming. The control of fruits was produced from organic farming while, the treated fruits were produced by conventional farming.
The collection of these fruits was done to preserve their initial micro flora. They were directly collected from the tree using gloves and kept in sterile bags on Oct. 2012. The collected samples were stored in a refrigerator then transferred via plane to Cirad Montpellier (France). The bacterial and yeast DNA were extracted from the fresh fruits. The farming types of the samples were defined as well as the harvest date.
2.2 Extraction of DNA from bacteria and yeast
This technique was carried out according to the protocol described by El Sheikha et al. [Citation7].
2.3 PCR-DGGE analysis
The specific amplification of the 16S rDNA was carried out via PCR using universal primers for the bacterial domain described by Muyzer et al. [Citation8]. Gc338f (5′CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGACTCCTACGGGAGGCAGCAG, Sigma Company, France) and 518r (5′ATTACCGCGGCTGCTGG, Sigma Company). The amplification of the V3 variable region of the 16S rDNA of peaches' bacterial communities was realized using GC clamp that consists of 40 nucleotides. The addition of clamp to forward primer 5′ of 338f was to ensure that DNA fragment will remain partially double stranded [Citation9]. Each PCR mixture is composed of about 100 ng of DNA template, the primers (0.2 µM), the dNTPs (200 µM), 5 µL MgCl2 (1.5 mM) of 10X of taq buffer MgCl2 free (Promega Company, France) and 5 units of the Tag polymerase enzyme (Promega Company). The initial denaturation was done at 95 °C for 1 min and ten touchdown cycles of denaturation at 95 °C for 1 min, followed by annealing at 65 °C for 1 min and finally extension at 72 °C for about 3 min followed by twenty cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 3 min.
Also, the amplification of D1/D2 region of the 26S rRNA gene was carried out via eukaryotic universal primers “NL1GC (5′-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCC ATA TCA ATA AGC GGA GGA AAA G-3′”, Sigma Company) and the reverse primer “LS2 (5′-ATT CCC AAA CAA CTC GAC TC-3′”, Sigma Company) amplifying a 250 base pair fragment [Citation7,Citation10]. A 30 bp GC-clamp (Sigma Company) was added to NL1. The PCR was done in a final volume of 50 µL containing 0.2 µM of each primers, dNTPs (200 µM), MgCl2 (1.5 mM), 5 µL of reaction Taq buffer MgCl2 free (10×) (Promega Company), 1.25 Unit of Taq DNA polymerase (Promega Company) and 2 µL of the extracted DNA {≈30 ng}. The PCR was run for about thirty cycles with annealing temperature at 52 °C for 2 min, extension at 72 °C for 2 min, and denaturation at 95 °C for 60 s [Citation7].
About 5 µL of the PCR products for bacteria and yeasts were analyzed initially using conventional electrophoresis 2% (w/v) agarose gel with TAE 1 × buffer, stained with ethidium bromide and quantified by a standard of DNA mass ladder 100 bp (Promega Company).
The PCR products were analyzed using DGGE via a Dcode™ universal mutation detection system (Bio-Rad Laboratories, USA) using the procedure first described by Muyzer et al. [Citation8] and modified by El Sheikha et al. [Citation7]. Samples were loaded into 8% (w/v) polyacrylamide gels (acrylamide/N,N′-methylene bisacrylamide, 37.5/1, Promega Company) in 1 × TAE buffer (40 mM Tris–HCl, pH 7.4, 20 mM sodium acetate, 1.0 mM Na2-EDTA).
All electrophoresis experiments were done at 60 °C using a denaturing gradient ranging from 30% to 60% (100% corresponded to 7 M urea and 40% [v/v] formamide, Promega Company). The gels were electrophoresed at twenty volts for ten minutes and then at eighty volts for twelve hours. After electrophoresis, the gels were stained with ethidium bromide for 30 min and then rinsed for ten minutes in distilled water and then photographed on a UV trans illuminator using the Gel Smart 7.3 system (Clara Vision, Les Ulis).
2.4 Statistical and image analysis
All of the gel images' Individual lanes were straightened and aligned via Image Quant TL – software, v.2003. This software allowed identifying the relative positions of bands.
The generated banding pattern in the analysis of DGGE is considered as a complete photo of all of the dominant bacteria and yeast in the populations. Any individual distinct band refers to a unique “sequence type” [Citation11]. This was ensured by Kowalchuk et al. [Citation12], who demonstrated that co-migrating bands were generally linked to an identical sequence. The PCR-DGGE fingerprints were scored manually via the presence and absence of co-migrating bands, regardless of intensity. Pair wise community similarities were quantified via the Dice similarity coefficient (SD) [Citation13].
where, Na refers to the band number recorded in A sample, Nb refers to the band number in B sample, and Nc refers to the band numbers that are common into both samples. The Similarity index was expressed ranging from 0 (complete dissimilarity) to 100 (perfect similarity). Valuable differences of bacteria and yeast communities of peach fruits from two farming types were determined via the factorial correspondence analysis (FCA) using the first two variances which described most of the variation in the whole data set.
2.5 DNA bands sequence analysis and microbial identification
The DGGE gel DNA bands were selected and excised carefully from the gel by means of sterile cutter. The gel pieces were soaked into TE buffer (100 mL) overnight (4 °C). Each band Eluted DNA was purified using Wizard PCR Preps kit DNA Purification system (Promega Company), and then the DNA purified was amplified again via the same PCR conditions as mentioned above by using primers (without GC-clamp). The PCR amplicons were sequenced via GATC Biotech (Germany). The analysis of DNA base sequences were performed via comparison with the databases of GenBank of NCBI. Then, searches in GenBank via BLAST program were done in order to detect the closest recorded relative of partial 16S and 26S rDNA sequences [Citation14].
3 Results
3.1 Fruits sampling
Mature peach fruits were collected in Jordan from two farming types which are organic farming and conventional farming (Fig. 1).
3.2 Extraction of bacterial and yeast DNA from Jordan peach fruits from two farming types and confirmation of PCR amplification of DNA extracted
The extraction of DNA of the bacterial and yeast communities that are present on peach fruits was verified on agarose gel {0.8% (w/v)} and achieved very good patterns.
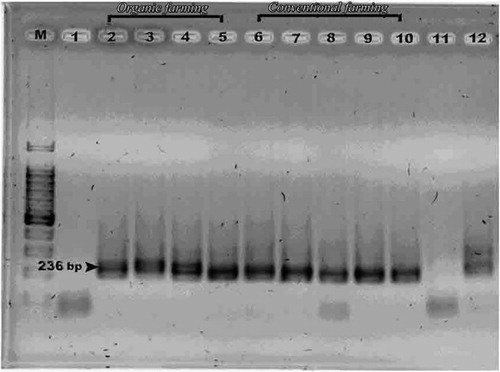
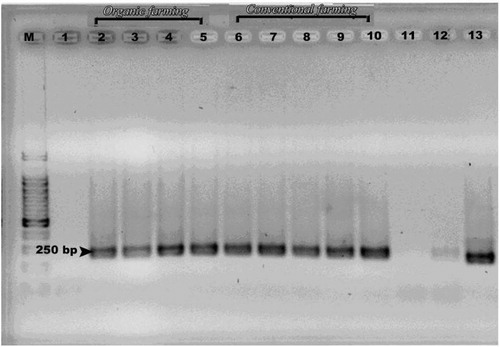
The amplification efficiency of the fractions was performed via electrophoresis of the PCR products (amplicons) on agarose gel {2% (w/v)} at 100 V for 30 min in TEA buffer. All resulted bands were clearly observed and had a molecular size of 250 base pairs (yeast) and 236 base pairs (bacteria) (Figs. 2 and 3). The band intensity of the PCR products (amplicons) was very important and confirmed that the amplification of bacterial and yeast DNA was done very well and by this way we can continue to make analysis of all of these amplicons via DGGE method.
3.3 Bacterial and yeast DNA. DGGE pattern from Jordan peach fruits via different farming types
The observed and obtained bands from DGGE gels had good and enough intensities that enable us to analyze bacteria and yeast DNA samples extracted from Jordan peach fruits (Fig. 4), so, the whole DNA quantity that was deposited in the DGGE gel was enough to use bacteria and yeast DNA as potent markers in order to detect Jordan peach fruits' farming type. Each of the vertical lines refers to a fruit farming type and each of the spots refers to a bacterial or yeast species. The replica of PCR-DGGE patterns of Jordan peach fruits for each farming type was the same for each type and indicated presence of 4 to 11 bands for each peach fruit farming type.
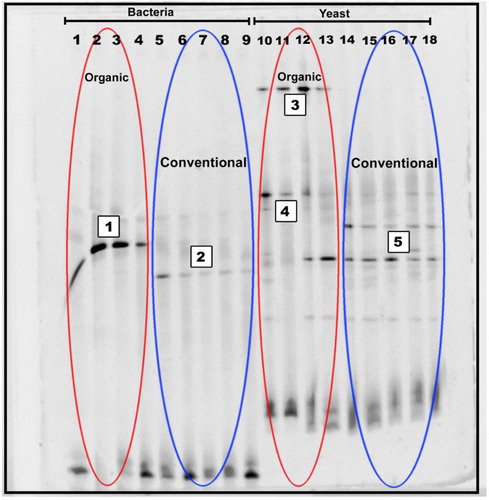
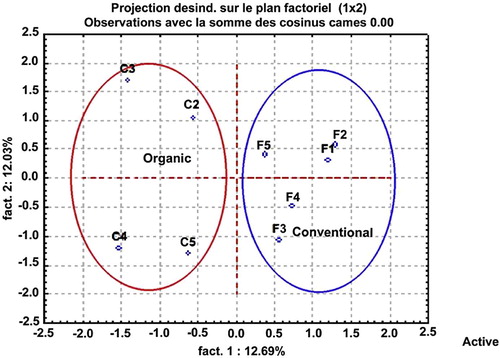
Furthermore, the Factorial Correspondence Analysis (FCA) was confirmed to be a potential statistical analysis tool for comparing the bacterial and yeast communities similarity of Jordan peach fruit samples according to farming types during the harvested season (Fig. 5). Finally, we can see clearly that the two different farming types were represented by two different groups.
3.4 Identification of dominant bacteria and yeasts on peach fruits from two different farming types via sequencing
The characterization of the bacterial and yeast population contaminated Jordan peach fruits were performed using molecular analysis via PCR DGGE and sequencing. By this way the dominant micro flora that was present in the samples can be detected and identified via sequencing of the gel interesting bands.
To our information, there is no knowledge on the bacterial and yeasts populations from these two different Jordan peach farming types that were obtained via cultural methods. Excised bands from DGGE gel of peach fruits extracted yeast and bacterial DNA were sequenced to identify bacterial and yeasts strains that present on the samples. Each band sequence refers to “a unique sequence” which is enough for identification via comparison in between the obtained sequences and BLAST standard references based upon the phylogenetic relations of the 236 and 250 base pairs of partial 16S rDNA and 26S rDNA sequences of hyper variable regions ().
Table 1 Sequencing results of the bands cut from the DGGE gel.
3.5 Microbial diversity of the excised DGGE bands
In order to describe changes in the bacterial and yeast populations that were present on farmed Jordan peach fruits from the two farming types, a molecular approach including PCR-DGGE and sequencing was performed. It reveals microbial diversity between the two farming types.
3.5.1 Identification of bacteria from their DNA
Sequence analyses of two bands, shown in Fig. 4, permitted to identify Leuconostoc pseudomesenteroides as the dominating bacteria in the organic peaches. This strain is traditionally found in association with plant matter, fruits etc. and could cause food spoilage [Citation15], which may appear in many diverse forms, all being consequences of the accumulative effects of spoilage compounds. Thus, the spoilage potential of leuconostoc is not solely dependent on their growth, but also on their metabolic activities. The spoilage metabolites produced reflect the physiological activities of the cell and hence vary depending on the conditions of the food system, involving oxygen tension, pH and carbon sources available.
The second band was identified as Lactobacillus coryniformis which is regarded as the main bacteria in conventional farming type of peach. This strain is one of the most important lactic acid bacteria. Lactic bacterial cluster name is due to the ability of bacteria to convert lactose into lactic acid. The production ability of lactic acid makes its media acidic that in turn inhibits growth of some other harmful or pathogenic bacteria and fungi and favors good plant growth and development [Citation16]. Furthermore, these bacteria may have some potent therapeutic properties (anti-inflammatory, anti-cancer activities etc.).
3.5.2 Identification of yeasts from their DNA
The three band sequence analyses that were included in the indicated bands on “Fig. 4”, ensured that two of them were identified as Pichia kluyveri and Pichia fermentans that have been reported to be part of the yeast micro flora in the organic farming type of peach. This genus of yeasts belongs to the family Saccharomycetaceae and most of these strains are usually found in decaying plants and fruit juices. The anamorphs of some Pichia species are Candida species. Some Pichia species have recently been clinically proven to be pathogens, well known as opportunistic pathogens in fruits [Citation17].
Finally, Tremella flava (TF) was detected as the predominant yeast in the conventional farming type of peach at the end of the trail. This strain is a genus of fungi of the family Tremellaceae and this species has anti-inflammation and anti-infection applicability [Citation18]. So, many researchers suggested that TF might be a novel, natural alternative for use as anti-inflammation and an anti-infection agent, so it is valuable for fruits in this farming case.
4 Discussion
Several scientists already used the PCR-DGGE technique in order to analyze the bacterial and yeast populations in fruits and their products [Citation7,Citation19–Citation[20]Citation21]. However, we think that our paper is one of the few publications that introduced the bacterial and yeast populations analysis in peach fruits to differentiate different farming types by PCR-DGGE.
In our study, we proved that the DGGE pattern of the bacterial and yeast populations DNA isolated from Jordan peach fruits were strictly linked to the treatments applied to the fruits. Analysis of peach fruits originated from two different farming types that showed some significant differences in the migration profiles on DGGE gel. However, the replica for each farming mode gave similar DGGE profiles through the research.
The differences in microbial environment resulted from differences in the band profiles that are greatly influenced by the farming types. The processing system types applied could also influence the microbial populations of peach fruits. Some common bacteria and yeast bands were observed in the DGGE gel in all of the peach fruit samples.
We saw also that there was a complete statistical linkage between the bacterial and yeast populations and the farming mode, if we compared by FCA for DGGE profile for different farming modes of fruits. We can conclude that there were sufficient environmental differences between the two different farming types where the Jordan peach fruits were harvested to have a major effect on the bacterial and yeast ecology. Based upon this, we could make a statistical relationship between the farming mode and the bacterial yeast populations.
Finally, analysis of peach fruit bacterial yeast populations via PCR-DGGE could be applied to discriminate different farming types. This global technique is very quick (i.e. <24 h) compared with all of the other classical and traditional microbial techniques used and avoids the biochemical analysis of microbes. In addition the molecular sizes of the bands permit the identification of strain via sequencing facility. By this way this technique can be suggested as a quick analytical traceability tool for fruits and could be considered as a supplier of “a unique molecular biological bar code” for each farming mode.
Acknowledgments
The authors thank Cirad, UMR 95 Qualisud, Montpellier, France and Egyptian Ministry of Higher Education for financial support of this scientific research work.
References
- M.FaustB.L.TimonOrigin and dissemination of peachHortic Rev3312010
- B.PeresN.BarletG.LoiseauD.MontetReview of the current methods of analytical traceability allowing determination of the origin of foodstuffsFood Control182007228235
- O.O.SodekoY.S.IzuagbeM.E.UkhunEffect of different preservative treatment on the microbial population of Nigerian orange juiceMicrobios511987133143
- D.MontetR.LeesingF.GemrotG.LoiseauDevelopment of an efficient method for bacterial diversity analysis: Denaturing Gradient Gel Electrophoresis (DGGE) In: Seminar on food safety and international trade, Bangkok, Thailand2004
- Y.HamdoucheT.GuehiN.DurandK.B.D.KedjeboD.MontetJ.-C.MeileDynamics of microbial ecology during cocoa fermentation and drying: towards the identification of molecular markersFood Control482015117122
- YangC.H.D.E.CrowleyJ.A.Menge16S rDNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophthora infected avocado rootsFEMS Microbiol Ecol352001129136
- A.F.El SheikhaA.CondurI.MétayerD.D.Le NguyenG.LoiseauD.MontetDetermination of fruit origin by using 26S rDNA fingerprinting of yeast communities by PCR-DGGE: preliminary application to Physalis fruits from EgyptYeast26102009567573
- G.MuyzerE.C.De WaalA.G.UitterlindenProfiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNAAppl Environ Microbiol591993695700
- V.C.SheffieldJ.S.BeckE.M.StoneR.M.MyersAttachment of 40bp G+C rich sequence (GC-clamp) to genomic DNA fragments by polymerase chain reaction results in improved detection of singlebase changesProc Natl Acad Sci USA861989232236
- L.CocolinL.F.BissonD.A.MillsDirect profiling of the yeast dynamics in wine fermentationsFEMS Microbiol Lett18920008188
- G.MuyzerA.TeskeC.O.WirsenH.W.JannaschPhylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent sample by denaturing gradient gel electrophoresis of 16S rDNA fragmentArch Microbiol1641995165172
- G.A.KowalchukJ.R.StephenW.De BoerJ.I.ProsserT.M.EmbleyJ.W.WoldendorpAnalysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR amplified 16S ribosomal DNA fragmentsAppl Environ Microbiol63199714891497
- M.HeyndrickxL.VauterinP.VandammeK.KerstersP.De VosApplicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomyJ Microbiol Methods261996247259
- S.F.AltschulT.L.MaddenA.A.SchafferZhangJ.ZhangZ.W.Milleret alGapped BLAST: a new generation of protein database search programsNucleic Acids Res25199733893402
- Y.HamasakiM.AyakiH.FuchuM.SugiyamaH.MoritaBehavior of psychrotrophic lactic acid bacteria isolated from spoiling cooked meat productsAppl Environ Microbiol69200336683671
- V.K.BatishU.RoyR.LalS.GroverAntifungal attributes of lactic acid bacteriaCrit Rev Biotechnol171997209225
- P.S.MamakosOutlines the production method of Birch derived Xylitol Birch-Xylitol Manufacturing Report2009
- ChenT.-I.I.-C.SheihH.-Y.Joann JengFangT.J.Anti-inflammation and anti-infection applicability of Tremella flava Chen fermented soymilk (TFS) in a BALB/c mice modelInt Proc Chem Biol Environ772014610
- C.J.PrakitchaiwattanaaG.H.FleetG.M.HeardaApplication and evaluation of denaturing gradient gel electrophoresis to analyze the yeast ecology of wine grapesFEMS Yeast Res42007865877
- A.F.El SheikhaI.MétayerD.MontetA biological bar-code for determining the geographical origin of fruit by using 28S rDNA fingerprinting of fungi communities by PCR-DGGE: an application to Physalis fruits from EgyptFood Biotechnol2522011115129 A.F.El SheikhaD.MontetDetermination of fruit origin by using 28S rDNA fingerprinting of fungal communities by PCR-DGGE: an application to Physalis fruits from Egypt, Uganda and ColombiaFruits66220117990
- A.F.El SheikhaN.DurandS.SareterJ.B.OkulloD.MontetStudy of the microbial discrimination of fruits by PCR-DGGE: application to the determination of the geographical origin of Physalis fruits from Colombia, Egypt, Uganda and MadagascarFood Control24201212 57–63