Abstract
Emerging Microbes & Infections (2012) 1, e38; doi:10.1038/emi.2012.35
There is an urgent need for new strategies to combat the spread of drug-resistant bacteria worldwide. This commentary concerns, namely bacterial protein phosphorylation as a promising target for novel antibacterials.
The constant increase of bacterial resistance to conventional antibiotics has indeed become a dramatic public health problem, critically requiring the discovery of innovative antibacterial drugs with new modes of action. In the past decades, this situation has been worsened by the considerably reduced investment of large pharmaceutical companies in the research and development of antibiotics.Citation1
Antibiotic resistance is acquired by chromosome mutation and/or integration of plasmids/transposons that carry resistance determinants by means of horizontal gene transfer.Citation2 The main mechanismsCitation3 through which resistance can develop are: (i) qualitative or quantitative modification of the target; (ii) enzymatic inactivation of antibiotics by hydrolysis or structural alteration; (iii) prevention of drug accumulation due to the impermeability of bacterial cell or increased efflux; and (iv) mutations in drug-activating enzymes.Citation4,Citation5,Citation6 Since different types of antibiotics have been frequently used simultaneously, several bacterial species have evolved toward multiresistance.
In the search for new antibacterials, different strategies have been explored. One of them has consisted in bringing incremental improvements to existing antibiotics by chemical modification, with, however, the risk for the corresponding derivatives being rapidly ineffective against the prevailing resistance mechanisms.Citation1,Citation7 This strategy has been reinforced by extensive efforts made to better understand the mode of action at the molecular level of already known antibiotics, namely by using efficient techniques of structure determination such as nuclear magnetic resonance (NMR) and X-ray crystallography. In addition, new chemical classes have appeared, either natural or synthetic, and novel molecules have been assayed for therapeutic potential.Citation8 The use of combination therapies involving treatment of infections by sets of drugs rather than individual drugs has also been considered.Citation9 Still, in most cases, those different antibiotics have turned out to inhibit in fact the same four classical targets: nucleic acid biosynthesis, protein biosynthesis, cell wall formation and folic acid biosynthesis.Citation8
From the mid-1990′s, the availability of a large number of complete bacterial genome sequences has provided an impressive tool to identify a variety of new putative targets.Citation10 The genome-based technologies and high-throughput screenings have generated a renewed interest in the search of novel antibiotics, especially in small biotechnology companies and academic centers. This concerns gram-negative, as well as gram-positive species, given the fact that there are less effective agents for treating gram-negative infections.Citation11 To cite a few, the new targets include fatty acid biosynthesis, lipoprotein biogenesis, efflux pumps, protein secretion, riboswitches and some specific antimicrobial peptides.Citation12 However, the results obtained to date indicate that minimal success has been met in converting theses targets into drugs, since none of them has reached advanced clinical development yet.Citation1,Citation7 Revisiting the choice of targets and screen designs, and the compound libraries chosen for screens should help in improving the efficiency of this approach.Citation13
Another target of special interest concerns bacterial protein phosphorylation by endogenous specific enzymes, which represents a promising way toward the discovery of non-conventional antibacterial drugs. This post-translational modification was long claimed to be restricted to eukaryotes until its occurrence was first demonstrated, simultaneously and independently, in Escherichia coliCitation14 and Salmonella typhimurium.Citation15 Since then, its existence has been described in a multitude of bacterial species and it is now considered an ubiquitous process in non-eukaryotic organisms.
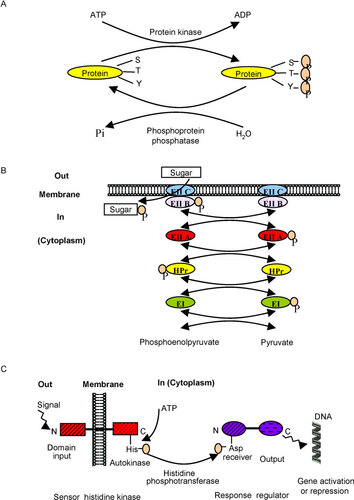
Whereas eukaryotes utilize basically only one type of phosphorylating machinery that operates through the modification of proteins at serine/threonine or tyrosine residues at the expense of adenosine triphosphate (ATP), bacteria possess a diversity of phosphorylating systems. One system is similar, but not identical, to that of eukaryotesCitation16,Citation17 (Figure 1A). Recent phosphoproteomics analyses have shown that bacterial serine/threonine and tyrosine kinases play vital roles in the molecular mechanisms of cell signaling and general regulation of cellular functions, such as central metabolism, cell growth, cell division and differentiation.Citation18 Therefore, these kinases and the cognate phosphoprotein phosphatases represent attractive antibacterial targets that deserve further investigation. Research in this field will obviously necessitate characterization of specific structural and functional features that differentiate the bacterial enzymes from their eukaryotic counterparts. On the other hand, bacteria are able to detect and transport different sugar substrates through their membrane, which are essential to their growth, by using the phosphoenolpyruvate: carbohydrate phosphotransferase system, which is a process strictly specific to prokaryotesCitation19 (Figure 1B). In this case, a phosphoryl group provided by phosphoenolpyruvate is passed down a cascade of five proteins or protein domains, and finally transferred to a sugar. Moreover, bacteria harbor a wide range of two-component systems (TCS) and their expanded variants known as phosphorelays to adapt to environmental conditions.Citation20 To do so, a sensor histidine protein kinase autophosphorylates in response to signal ligands and subsequently transfers its phosphate to an invariant aspartate residue of a response regulator that is generally a transcription factor (Figure 1C). Here again, since TCS are absent in higher eukaryotes, they can be selectively targeted by antibacterial molecules.
Of particular importance is the fact that the activity of these three major phosphorylating systems, which are not mutually exclusive, is closely connected with the virulence of pathogens.Citation21 Thus, the possibility to attenuate virulence by acting on this relationship represents a notably interesting approach to prevent the expression of pathogenicity without causing the arrest of growth or death of bacteria, as do conventional antibiotics. The ensuing advantage is the preservation of the host endogenous normal flora and the limitation of the selective pressure, which results in a decreased capacity of drug resistance.
To date, a number of antibacterial agents have already proven to impair virulence, although to a varying extent, by interfering with phosphorylation. For instance, the low-molecular-weight inhibitor AX20017 blocks selectively the activity of the serine/threonine protein kinase PknG, a virulence factor of Mycobacterium tuberculosis, by interacting with a unique set of amino-acid side chains in the kinase domain that are not found in any human kinase.Citation22 Also, the production and transport of bacterial surface polysaccharides that are potent virulence factors controlled by the activity of the tyrosine kinases called BY-kinases, are inhibited by peptide nucleic acid analogues able to bind specifically to the ATP-binding pocket of these enzymes.Citation23 In both phosphotransferase system and TCS systems, a variety of molecules stemming from library screens and structure–activity relationship programs have been checked for potential inhibition. But, so far, most of them have been shown not to be selective enough or even to have sometimes unexpected side effects.Citation24 However, in TCS systems, a small molecule of the benzenesulfonamide family, LED 209, has recently been shown to inhibit the binding of signals to the histidine protein kinase QseC, which prevents its autophosphorylation and consequently the virulence gene expression mediated by this kinase in several pathogens.Citation25 In a similar way, the infection process by Streptococcus pneumoniae is impaired by a series of chemical compounds, namely, furan and thiophene derivatives, which inhibit the histidine protein kinase VicK.Citation26 In general, the blockade of histidine kinase activity is not lethal to the bacterial cell. Nevertheles, the deranged regulation that occurs consequently results in a bacteriostatic effect that can be sufficient to hinder infection. This applies as well to serine/threonine and tyrosine kinases. Thus, for example, in-frame deletions of the stk1 gene that encodes serine/threonine kinase Stk1 in Staphylococcus aureus cause a strong reduction of bacterial growth in mouse kidneys compared to the parental strain.Citation27
The concept that protein phosphorylation in its various facets could be a good drug target is not new per se. However, the data summarized here, as well as other similar reports, confirm that further investigation of this protein modification raises hope for the future discovery of novel antibacterials. Obviously, a number of technical questions will have to first be answered such as the availability of drugs with broad spectrum activity or the immediacy of their action in clinical use. However, the combination of bacterial genomics, biochemistry coupled with bioinformatics, and physiology can be expected to facilitate valuable progress in this field. In particular, the recent determination of the intimate three-dimensional structure of a few serine/threonine kinases (reviewed in Ref. 16) and tyrosine kinases,Citation28,Citation29,Citation30 and more to come, should offer significant opportunities for designing previously unexploited molecules that would efficiently combat bacterial diseases by acting on this protein modification.
- Jabes D. The antibiotic R&D pipeline: an update. Curr Opin Microbiol2011; 14: 564–569.
- Schultz C, Geerlings S. Plasmid-mediated resistance in Enterobacteriaceae: changing landscape and implications for therapy. Drugs2012; 72: 1–16.
- Martinez M, Silley P. Antimicrobial drug resistance. Handb Exp Pharmacol2010; 199: 227–264.
- Zhang Y, Heym B, Allen B et al. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature1992; 358: 591–593.
- Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, causes resistance to antituberculous drug pyrazinamide in tubercle Bacillus. Nature Medicine1996; 2: 662–667.
- Goodwin A, Kersulyte D, Sisson G et al. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol1998; 28: 383–393.
- Theuretzbacher U. Resistance drives antibacterial drug development. Curr Opin Pharmacol2011; 11: 433–438.
- Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nature Rev Microbiol2010; 8: 423–435.
- Fischbach MA. Combination therapies for combating antimicrobial resistance. Curr Opin Microbiol2011; 14: 519–523.
- Roemer T, Davies J, Giaever G et al. Bugs, drugs and chemical genomics. Nat Chem Biol2011; 8: 46–56.
- Bassetti M, Ginocchio F, Mikulska M et al. Will new antimicrobials overcome resistance among Gram-negatives? Expert Rev Anti Infect Ther2011; 9: 909–922.
- Ma Q, Yu Z, Han B et al. [Research progress in fusion expression of antimicrobial peptides.] Sheng Wu Gong Cheng Xue Bao2011; 27: 1408–1416. Chinese.
- Pucci MJ. Novel genetic techniques and approaches in the microbial genomics era: identification and/or validation of targets for the discovery of new antibacterial agents. Drugs RD2007; 8: 201–212.
- Manai M, Cozzone AJ. Analysis of the protein kinase activity of Escherichia coli cells. Biochem Biophys Res Commun1979; 91: 819–826.
- Wang JH, Koshland DE Jr. Evidence for protein kinase activities in the prokaryote Salmonella typhimurium. J Biol Chem1978; 253: 7605–7608.
- Pereira SFF, Goss L, Dworkin J. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Molec Bio Rev2011; 75: 192–212.
- Grangeasse C, Cozzone AJ, Deutscher J et al. Tyrosine phosphorylation: an emerging device of bacterial physiology. Trends Biochem Sci2007; 32: 86–94.
- Mijakovic I, Macek B. Impact of phosphoproteomics on studies of bacterial physiology. FEMS Microbiol Rev2012; 36: 877–892.
- Lengeler JW, Jahreis K. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib Microbiol2009; 16: 65–87.
- Goulian M. Two-component signaling circuit structure and properties. Curr Opin Microbiol2010; 13: 184–189.
- Ge R, Shan W. Bacterial phosphoproteomic analysis reveals the correlation between protein phosphorylation and bacterial pathogenicity. Genom Proteom Bioinform2011; 9: 119–127.
- Scherr N, Honnappa S, Kunz G et al. Structural basis for the specific inhibition of protein kinase G, a virulence factor of Mycobacterium tuberculosis. Proc Natl Acad Sci USA2007; 104: 12151–12156.
- Grangeasse C, Nessler S, Morera S et al. Inhibitors of bacterial tyrosine kinase and uses thereof. World Intellectual Property Organization. WO/2009/133209A1, 2009
- Stephenson K, Hoch JA. Two-component and phosphorelay signal-transduction systems as therapeutic targets. Curr Opin Pharmacol2002; 2: 507–512.
- Rasko DA, Moreira CG, Li DR et al. Targeting QseC signaling and virulence for antibiotic development. Science2008; 321: 1078–1080.
- Li N, Wang F, Niu S et al. Discovery of novel inhibitors of Streptococcus pneumoniae based on the virtual screening with the homology-modeled structure of histidine kinase VicK. BMC Microbiol2009; 9: 129–139.
- Debarbouillé M, Dramsi S, Dussurget O et al. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol2009; 191: 4070–4081.
- Olivares-Illana V, Meyer P, Bechet E et al. Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLoS Biol2008; 6: e143.
- Lee DC, Zheng J, She YM et al. Structure of Escherichia coli tyrosine kinase Etk reveals a novel activation mechanism. EMBO J2008; 27: 1758–1766.
- Bechet E, Gruszczyk J, Terreux R et al. Identification of structural and molecular determinants of the tyrosine kinase Wzc and implications in capsular polysaccharide export. Molec Microbiol2010; 77: 1315–1325.