ABSTRACT
Capsule
Analysis of mitochondrial DNA from reed warblers sampled in Italy reveals that they are most similar to the nominate race of European Reed Warbler Acrocephalus scirpaceus scirpaceus.
Aim
To fill gaps in our knowledge of the distribution of European/African Reed Warblers Acrocephalus scirpaceus/baeticatus in the Mediterranean region through genetic analysis of populations from the key biogeographic areas of Sicily, Sardinia and mainland Italy.
Methods
We assessed the genetic placement of Italian reed warbler populations within a comprehensive dataset of this species complex, by sampling these birds from principal peninsular and insular breeding populations (for a total of 17 samples) and by comparing their mitochondrial cyt b sequences with those available in GenBank-NCBI.
Results
The final dataset included 171 cyt b sequences (763 base pair long). The analysis showed samples from Italy had a high similarity with the European Reed Warbler A. s. scirpaceus subspecies. This fills an important information gap in the phylogeography of the group.
Conclusion
The genetic cohesion shown between Italian and other European populations may denote a structuring in at least three distinct refugia in the Western Palearctic: Iberia for A. baeticatus ambiguus, the Caucasus Mountains for A. s. fuscus, and both Italy and the Balkans for A. s. scirpaceus.
The Mediterranean basin is ranked among the 25 global biodiversity hotspots (Myers et al. Citation2000). The higher conservation priority of this area is due both to its taxonomic richness and uniqueness, and to anthropogenic threats causing a severe loss of habitat (Cuttelod et al. Citation2008). Responses to paleo-climatic events, such as isolation in refugial areas during glacial periods, are documented as one of the drivers that led to the high phylogenetic diversity in this region (Bilton et al. Citation1998, Stewart et al. Citation2010, Voelker and Light Citation2011) and to the frequent occurrence of cryptic species in a wide range of taxa, including marine organisms (Calvo et al. Citation2009), freshwater arthropods (Bernini et al. Citation2016), amphibians (Carranza et al. Citation2008), reptiles (Ghielmi et al. Citation2016), mammals (Hulva et al. Citation2004) and birds (Brambilla et al. Citation2008, Pellegrino et al. Citation2017). Genetic tools have played a prominent role in our understanding of these processes and in assessing possible taxonomic delimitation, the implications of which extend into the fields of biogeography and conservation (Lohman et al. Citation2010, Pertoldi et al. Citation2012, Galimberti et al. Citation2021).
Along with Iberia and the Balkans, the Italian peninsula and its islands represent a well-documented Pleistocene glacial refugium, which facilitated an evolutionary radiation area for many taxa, including several bird species (Brito Citation2007, Pellegrino et al. Citation2015, Drovetski et al. Citation2018). In birds, isolation dynamics and the subsequent evolution of unique genotypes within the Italian refugium are generally more common for sedentary or near-sedentary species (Förschler et al. Citation2009, Pellegrino et al. Citation2016). However, these divergence mechanisms also involve some long-distance migratory bird species, for which the Italian peninsula and the western Mediterranean islands have been a cradle of genetic isolation processes (Brambilla et al. Citation2008, Pons et al. Citation2016, Citation2019, Zuccon et al. Citation2020).
Among long-distance migrant species breeding extensively (but not necessarily exclusively) around the Mediterranean, the European/African Reed Warbler Acrocephalus scirpaceus/baeticatus complex has attracted the attention of many researchers, who have investigated, through molecular and morphological methods, the species/subspecies boundaries and evolutionary relationships of the group (Procházka et al. Citation2011, Arbabi et al. Citation2014, Hering et al. Citation2016, Olsson et al. Citation2016, Pavia et al. Citation2018, Babbington et al. Citation2019). According to the International Ornithological Congress (Gill et al. Citation2020), this species complex is considered to be composed of two species: the European Reed Warbler Acrocephalus scirpaceus with four subspecies (A. s. scirpaceus, A. s. fuscus, A. s. avicenniae, and A. s. ammon), and the African Reed Warbler Acrocephalus baeticatus with six subspecies (A. b. baeticatus, A. b. ambiguus, A. b. minor, A. b. cinnamomeus, A. b. suahelicus, and A. b. hallae). This assessment is mostly in accordance with the mitochondrial cytochrome b analysis presented by Olsson et al. (Citation2016), wherein they proposed to treat the complex as one polytypic species (A. scirpaceus) with seven well-supported clades divided in two main clusters: a Eurasian group that included the subspecies Acrocephalus scirpaceus scirpaceus, A. s. fuscus and A. s. avicenniae, and an African group encompassing not only the Sub-Saharan subspecies A. s. baeticatus, A. s. minor, A. s. hallae, A. s. cinnamomeus, but also the Iberian and Moroccan populations. These latter populations belong to the resurrected taxon A. scirpaceus ambiguus, which is characterized by intermediate morphometries between A. s. scirpaceus and A. s. baeticatus (Olsson et al. Citation2016).
While the taxonomy and evolutionary history of the complex are becoming clearer, there are several aspects still in need of further study. One important aspect is increased sampling, as genetic information from key biogeographic regions (e.g. the Italian peninsula, Sardinia and Sicily) is still missing. Here, we aim to assess the genetic relationships of Italian populations of reed warblers relative to other populations, by adding new genetic data to that of Olsson et al. (Citation2016). We specifically seek to address several questions. First, are some or all of the Italian populations genetically differentiated from non-Italian populations? We hypothesize that the population most likely to be differentiated is the Sardinian one, where numerous endemic bird taxa (Giglio-Tos Citation1918) and unique endoparasite lineages (Pellegrino et al. Citation2021) have been described. Second, if some or all of the Italian populations are not distinct, to which other populations are they most closely related? In this case we are unable to make predictions, as Italian bird populations have variably been most closely related to populations in Spain and France (Hourlay et al. Citation2008), and the Balkans and Europe more broadly (Drovetski et al. Citation2018, Raković et al. Citation2019). Further, while connections to northern African populations are possible, and have been observed in other taxa, including butterflies (Habel et al. Citation2009), shrews (Cosson et al. Citation2005) and frogs (Stöck et al. Citation2008), such a link has not as yet been established for populations of Italian avifauna. Nevertheless, several bird taxa endemic to the Western Palearctic include distributions both on Sardinia and Sicily (e.g. Spotless Starling Sturnus unicolor and Purple Swamphen Porphyrio porphyrio; Keller et al. Citation2020), as well as northwest Africa, suggesting a possible biogeographic connection between these southwestern Mediterranean areas.
Considering that (i) the Mediterranean and Italian areas have a well-established biogeographic potential as a diversity incubator, even for long-distance migrant birds; (ii) there is a lack of genetic assessment of reed warblers of some Mediterranean areas, such as Italy and its islands, (iii) taxonomic studies show genetic affinities between Italy and other European refugia, as well as northern Africa, we aim to assess the genetic affinities of Italian reed warbler populations by analysing the most comprehensive genetic dataset of the Eurasian/African species complex to date.
Methods
Sampling, DNA amplification and sequencing
Sampling of Reed Warblers took place in four geographic areas (i.e. northern Italy, southern Italy, Sardinia and Sicily), which generally correspond to areas of intraspecific genetic divergence previously identified in a wide range of taxa, such as in dragonflies and damselflies (Galimberti et al. Citation2021), butterflies (Scalercio et al. Citation2020), amphibians (Mattoccia et al. Citation2005), reptiles (Kindler and Fritz Citation2018), mammals (Wauters et al. Citation2017) and birds (Pellegrino et al. Citation2015, Zuccon et al. Citation2020) (see for sampling details). Birds were trapped using mist-nets set in Common Reed Phragmites australis dominated wetlands. Blood was drawn via brachial venipuncture using a small needle to puncture the vein, and a capillary tube to collect the blood. Blood was stored in 2 ml tubes filled with Queen's lysis buffer and samples were vouchered and placed in the collection of the University of Eastern Piedmont, Italy. For molecular analyses we included a subsample of 17 breeding individuals, as determined via the presence of a cloacal protuberance or brood patch (Svensson Citation1992) ().
Table 1. Sampling details of reed warbler specimens collected from Italy for this study. The identifier of sampling sites in the map of , the cyt b haplotypes and the GenBank accession numbers of the sequenced samples are also reported (* haplotypes discovered in this study).
DNA was extracted from approximately 25 to 50 µL of blood using the E.Z.N.A. Tissue DNA extraction kit (Omega Bio-Tek, Norcross, GA) following the manufacturer’s instructions. A 1022 base pair (bp) region of the mitochondrial cytochrome b (cyt b) gene was amplified using the primer pair L-14995/H-16065 (Helbig et al. Citation1995). Polymerase chain reaction (PCR) conditions consisted of an initial denaturation step at 94 °C for 5 min, followed by 35 cycles at 94 °C for 1 min, 52 °C for 1 min and 72 °C for 1 min, with a final extension step at 72 °C for 10 min. Sequencing was performed bi-directionally at Eurofins Genomics (Milan, Italy) using the same PCR primers. Consensus sequences were obtained by editing the electropherograms with Bioedit 7.2 (Hall Citation1999). After primer trimming, the presence of an open reading frame was verified for the obtained consensus sequences by using the on-line tool EMBOSS Transeq (http://www.ebi.ac.uk/Tools/st/emboss_transeq/). This analysis was conducted to exclude the amplification of pseudogenes that could affect identification results or the discovery of divergent intraspecific lineages. Consensus sequences were deposited in EMBL GenBank (accession numbers are listed in ).
Dataset assembly and haplotype structure analysis
Orthologous cyt b sequences, belonging to reed warbler subspecies breeding in the Western Palearctic (i.e. A. s. scirpaceus, A. s. fuscus, A. s. avicenniae, and A. b. ambiguus) were downloaded from GenBank-NCBI; no sequences were available for A. s. ammon (Hering et al. Citation2016). Moreover, we also considered reference cyt b sequences of A. baeticatus subspecies breeding in sub-Saharan Africa (i.e. A. b. baeticatus, A. b. minor, A. b. hallae and A. b. cinnamomeus), to calculate their genetic distance values from Western Palearctic subspecies and verify their possible genetic affinity to the Italian populations. Sequences showing insertions/deletions, that were missing more than 1% of sites or that were overlapping less than 750 bp with the region amplified and sequenced for the Italian samples, were discarded. Overall, a total of 154 cyt b sequences were retrieved from the public repository and combined with the 17 sequenced Italian reed warblers to constitute a unique dataset (see online Table S1).
Sequences were aligned with MAFFT 7.110 (Katoh and Standley Citation2013) using the E-INS-I option and cut to a final alignment length of 763 bp. This length was a compromise between including the highest number of taxa in the European/African Reed Warbler complexes and conserving the highest nucleotide information. As no phylogenetic analyses were necessary for the purpose of this study, we did not estimate the best-fit model for the dataset. However, to provide a measure of genetic distance among the Italian populations and the other subspecies known for the Mediterranean and the African Reed Warbler taxa, we calculated average uncorrected pairwise genetic distances (p-distance) using MEGA X (Kumar et al. Citation2018). The average within-group genetic distances (inferred with MEGA X) for each subspecies and country of origin were also calculated (online Table S2).
Identical sequences were collapsed into unique haplotypes using FaBox 1.5 online (Villesen Citation2007) (Table S1). In order to investigate the haplogroup assignment of Italian reed warbler cyt b haplotypes, a Median Joining network, encompassing all the Italian and Palearctic populations (, Table S1), was built using the median-joining algorithm implemented in PopART 1.7 (Leigh and Bryant Citation2015).
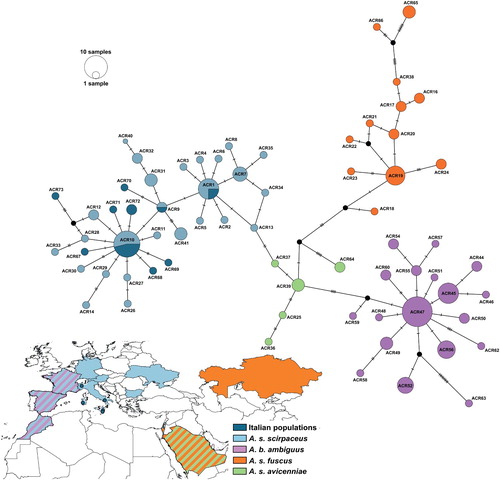
Figure 1. Median-joining network of cyt b haplotypes of Western Palearctic Acrocephalus scirpaceus/A. baeticatus species complex populations (, Table S1). Each circle represents a haplotype and circle size is proportional to haplotype frequency. Colours indicate different subspecies and blue dots also account for Italian sampling sites. Dashes represent substitutions. Distribution map is shown on the countries boundaries basis.
Results
We obtained high-quality full-length cyt b sequences for all 17 Italian samples. No sequences contained insertions or deletions, stop codons, or were biased by nuclear mitochondrial DNA interference. The comprehensive cyt b dataset assembled in this study included 171 sequences 763 bp long, which were collectively assigned to 82 haplotypes, 10 of which were found in reed warblers from Italy (, , Appendix Table S1). Of the 10 haplotypes found in Italian reed warblers, seven were exclusive to the country: three were found in Sardinia (ACR67, ACR72, ACR73), two in Sicily (ACR70, ACR71) and two in Southern Italy (ACR68, ACR69) (). The other sequences from Italian individuals coincide with three previously described haplotypes: ACR1, sampled in Sardinia and Southern Italy, was also observed in France and Israel; ACR9, known from Morocco, was shared by a Sicilian individual; and ACR10, the most common haplotype in our populations (Sardinia, Sicily and Northern Italy), is widely distributed, having been found in the Canary Islands, Spain, Germany, France and Israel (Olsson et al. Citation2016) ().
Our Median Joining haplotypes network (, Figure S1) shows that the different subspecies of the European Reed Warbler complex are distinct from the African Reed Warbler subspecies, as they have no shared haplotypes. On the whole, all the Italian haplotypes fall in the haplogroups representing A. scirpaceus scirpaceus ().
The genetic distance (p-distances) calculated between our Italian samples and other sequences of A. s. scirpaceus (0.0039 ± 0.0011) was lower than those obtained between Italian samples and the other taxa (A. s. avicenniae, A. s. fuscus, A. b. ambiguus, A. b. baeticatus, A. b. minor, and A. b. hallae), which ranged from 0.0122 ± 0.0033 and 0.0261 ± 0.0052 (). For intragroup genetic distances, the Italian population showed a value falling in the range of other A. s. scirpaceus populations (0.0029 ± 0.0008; range 0 ± 0–0.0066 ± 0.0030) (Table S2).
Table 2. Between groups genetic divergence values of cyt b sequences, based on the uncorrected p-distance calculation (bottom left), for the taxa of European and African Reed Warblers considered in this study. Standard error values are reported above the diagonal.
Discussion
The placement of Italian samples, from northern and southern populations as well as from the two major islands of Sicily and Sardinia, indicates that all belong to the A. scirpaceus nominal subspecies, the European Reed Warbler taxon that occurs throughout Europe, with the exception of most of the Iberian Peninsula (Olsson et al. Citation2016). This result fills a significant gap in the sampling framework of reed warbler phylogeography in the Mediterranean basin, as the Italian reed warbler populations have not been included prior to this new study (Hering et al. Citation2016, Olsson et al. Citation2016, Babbington et al. Citation2019).
The lack of unique haplotype clustering of our samples relative to other reed warbler samples () supports a previous study based on microsatellite markers (Procházka et al. Citation2011) that revealed high levels of gene flow among A. s. scirpaceus populations. Despite the occurrence of seven unique haplotypes from Sicilian, Sardinian and southern Italian individuals, our findings do not support the co-occurrence of divergent lineages typical of other European/African Reed Warbler subspecies in Italy. Furthermore, while the ACR9 haplotype is shared only by a single Sicilian (this study) and a single Moroccan individual (sampled in Olsson et al. Citation2016), this relationship suggests a possible similarity in A. s. scirpaceus populations from the southern Mediterranean basin. However, a greater sampling effort of reed warblers from northwest Africa will be necessary to establish this pattern.
On the whole, based on the reliability of cyt b in characterizing the principal clades of this passerine group (Olsson et al. Citation2016), genetic cohesion between Italian reed warblers and other Central European individuals belonging to A. s. scirpaceus populations is evident. On the other hand, the distance between Italian individuals and A. s. fuscus is similar to that calculated between Italian A. s. scirpaceus and Iberian A. baeticatus ambiguus. This phylogeographic pattern (i.e. the genetic homogeneity occurring among the Italian peninsula, the Balkans and the rest of Europe, vs. the divergent haplogroups shown by Iberian and Caucasus populations) is also known from studies of other passerine species (e.g. Dunnock Prunella modularis, Drovetski et al. Citation2018 and Common Chiffchaff Phylloscopus collybita, Raković et al. Citation2019), and may denote a population structuring of Acrocephalus scirpaceus through distinct glacial refugia within the Western Palearctic: A. s. fuscus and A. s. avicenniae in refugia in Saudi Arabia and the south Caucasus, and A. s. scirpaceus in refugia in the Italian and Balkan peninsulas.
The presence of two distinct clades of reed warblers in the Iberian Peninsula, with a clear genetic delimitation between this area and the rest of Europe, is comparable with that observed in Savi's Warbler Locustella luscinioides, another long-distance migrant associated with wetland habitats (Neto et al. Citation2012). This convergence may indicate an important role of Iberia (and possibly of northwest Africa) as a disconnected refugium from the rest of the Mediterranean region for wetland passerines. Our results likely exclude the occurrence of African Reed Warbler haplotypes from the geographic areas we sampled in Italy, Sardinia and Sicily.
Interestingly, among European Reed Warblers from Sardinia, there are reported cases of individuals showing complete post-breeding moult (Nissardi and Zucca Citation2001), as well as of individuals wintering on the island (Nissardi Citation1998). These patterns of complete post-breeding moult and overwintering in the Palearctic also occur in African Reed Warbler A. b. ambiguus, and involve populations or individuals from both Morocco and Spain (Amezian et al. Citation2010, Nieto et al. Citation2018). However, Nieto et al. (Citation2018) has suggested over the past few decades that European Reed Warbler individuals belonging to different populations are overwintering in Iberia, and that this pattern is related to increasingly warmer winters. In the light of the genetic assessment presented here, such modifications of trait phenology are likely not exclusive to a specific haplogroup, but could be due to the influence of shared environmental and climatic factors within the Mediterranean basin that deserve to be further addressed.
While this study adds an important piece to the complex puzzle of the genetic structure and taxonomy of the reed warbler group, further studies are still needed to disentangle remaining biogeographic knowledge gaps. In particular, sampling from transition areas between the different taxa in Turkey and the Near East, and from isolated populations (e.g. Cyprus, Crete, northeastern Africa) would be valuable; data from these regions are currently lacking.
Supplemental Material
Download MS Word (249.2 KB)Acknowledgments
We are grateful to Davide Magnani for laboratory support. This is publication number 1642 of the Biodiversity Research and Teaching Collections at Texas A&M University and publication number 11 of the W.I.N.E.S. collaborative research group.
References
- Amezian, M., Cortes, J., Thompson, I., Bensusan, K., Perez, C., Louah, A., Aziz El Agbani, M. & Qninba, A. 2010. Complete moult of an undescribed resident taxon of the Reed Warbler Acrocephalus scirpaceus/baeticatus complex in the Smir marshes, Northern Morocco. Ardea 98: 225–234.
- Arbabi, T., Gonzalez, J., Witt, H.H., Klein, R. & Wink, M. 2014. Mitochondrial phylogeography of the Eurasian Reed Warbler Acrocephalus scirpaceus and the first genetic record of A. s. fuscus in Central Europe. Ibis 156: 799–811.
- Babbington, J., Boland, C., Kirwan, G.M., Alsuhaibany, A., Shirihai, H. & Schweizer, M. 2019. Confirmation of Acrocephalus scirpaceus avicenniae (Aves: Acrocephalidae) from mangroves on the Red Sea coast near Jazan, southwest Saudi Arabia. Zool. Middle East 65: 201–207.
- Bernini, G., Bellati, A., Pellegrino, I., Negri, A., Ghia, D., Fea, G., Sacchi, R., Nardi, P.A., Fasola, M. & Galeotti, P. 2016. Complexity of biogeographic pattern in the endangered crayfish Austropotamobius italicus in northern Italy: molecular insights of conservation concern. Cons. Genetics 17: 141–154.
- Bilton, D.T., Mirol, P.M., Mascheretti, S., Fredga, K., Zima, J. & Searle, J.B. 1998. Mediterranean Europe as an area of endemism for small mammals rather than a source for northwards postglacial colonization. P. R. Soc. Lond. B. Biol. 265: 1219–1226.
- Brambilla, M., Vitulano, S., Spina, F., Baccetti, N., Gargallo, G., Fabbri, E., Guidali, F. & Randi, E. 2008. A molecular phylogeny of the Sylvia cantillans complex: Cryptic species within the Mediterranean basin. Mol. Phylogenet. Evol. 48: 461–472.
- Brito, P.H. 2007. Contrasting patterns of mitochondrial and microsatellite genetic structure among Western European populations of Tawny Owls (Strix aluco). Mol. Ecol. 16: 3423–3437.
- Calvo, M., Templado, J., Oliverio, M. & Machordom, A. 2009. Hidden Mediterranean biodiversity: molecular evidence for a cryptic species complex within the reef building vermetid gastropod Dendropoma petraeum (Mollusca: Caenogastropoda). Biol. J. Linn. Soc. 96: 898–912.
- Carranza, S., Romano, A., Arnold, E.N. & Sotgiu, G. 2008. Biogeography and evolution of European cave salamanders, Hydromantes (Urodela: Plethodontidae), inferred from mtDNA sequences. J. Biogeol. 35: 724–738.
- Cosson, J.F., Hutterer, R., Libois, R., Sara, M., Taberlet, P. & Vogel, P. 2005. Phylogeographical footprints of the Strait of Gibraltar and Quaternary climatic fluctuations in the western Mediterranean: a case study with the Greater White-toothed Shrew, Crocidura russula (Mammalia: Soricidae). Mol. Ecol. 14: 1151–1162.
- Cuttelod, A., García, N., Abdul Malak, D., Temple, H. & Katariya, V. 2008. The Mediterranean: a biodiversity hotspot under threat. In J.-C. Vié, C. Hilton-Taylor & S. N. Stuart. (ed) The 2008 Review of the IUCN red List of Threatened Species, 89–101. IUCN Gland.
- Drovetski, S.V., Fadeev, I.V., Raković, M., Lopes, R.J., Boano, G., Pavia, M., Koblik, E.A., Lohman, Y.V., Red'kin, Y.A., Aghayan, S.A., Reis, S., Drovetskaya, S.S. & Voelker, G. 2018. A test of the European Pleistocene refugial paradigm, using a Western Palaearctic endemic bird species. P. R. Soc. B-Biol. Sci. 285: 0181606.
- Förschler, M.I., Senar, J.C., Perret, P. & Björklund, M. 2009. The species status of the Corsican Finch Carduelis corsicana assessed by three genetic markers with different rates of evolution. Mol. Phylogenet. Evol. 52: 234–240.
- Galimberti, A., Assandri, G., Maggioni, D., Ramazzotti, F., Baroni, D., Bazzi, G., Chiandetti, I., Corso, A., Ferri, V., Galuppi, M., Ilahiane, L., La Porta, G., Laddaga, L., Landi, F., Mastropasqua, F., Ramellini, S., Santinelli, R., Soldato, G., Surdo, S. & Casiraghi, M. 2021. Italian odonates in the Pandora's box: A comprehensive DNA barcoding inventory shows taxonomic warnings at the Holarctic scale. Mol. Ecol. Resour. 21: 183–200.
- Ghielmi, S., Menegon, M., Marsden, S.J., Laddaga, L. & Ursenbacher, S. 2016. A new vertebrate for Europe: the discovery of a range-restricted relict viper in the western Italian Alps. J. Zool. Syst. Evol. Res. 54: 161–173.
- Giglio-Tos, E. 1918. In A. Musso. (ed) Gli uccelli d’Italia. Manuale pratico per la determinazione delle specie italiane di uccelli con nozioni per la preparazione delle pelli. Torino, Italy.
- Gill, F., Donsker, D. & Rasmussen, P. 2020. IOC World Bird List (v10.2). doi:10.14344/IOC.ML.10.2.
- Habel, J.C., Dieker, P. & Schmitt, T. 2009. Biogeographical connections between the Maghreb and the Mediterranean peninsulas of southern Europe. Biol. J. Linn. Soc. 98: 693–703.
- Hall, A. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid. S 41: 95–98.
- Helbig, A.J., Seibold, I., Martens, J. & Wink, M. 1995. Genetic differentiation and phylogenetic relationships of Bonelli's Warbler Phylloscopus bonelli and Green Warbler P. nitidus. J. Avian Biol. 26: 139–153.
- Hering, J., Winkler, H. & Steinheimer, F.D. 2016. A new subspecies of Eurasian Reed Warbler Acrocephalus scirpaceus in Egypt. Bull. B O C 136: 101–128.
- Hourlay, F., Libois, R., D’Amico, F., Sarà, M., O’Halloran, J. & Michaux, J.R. 2008. Evidence of a highly complex phylogeographic structure on a specialist river bird species, the Dipper (Cinclus cinclus). Mol. Phylogenet. Evol. 49: 435–444.
- Hulva, P., Horáček, I., Strelkov, P.P. & Benda, P. 2004. Molecular architecture of Pipistrellus pipistrellus/Pipistrellus pygmaeus complex (Chiroptera: Vespertilionidae): further cryptic species and Mediterranean origin of the divergence. Mol. Phylogenet. Evol. 32: 1023–1035.
- Katoh, K. & Standley, D.M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780.
- Keller, V., Herrando, S., Voříšek, P., Franch, M., Kipson, M., Milanesi, P., Martí, D., Anton, M., Klvaňová, A., Kalyakin, M.V., Bauer, H.-G. & Foppen, R.P.B. 2020. European Breeding Bird Atlas 2: Distribution, abundance and change. European Bird Census Council & Lynx Edicions, Barcelona.
- Kindler, C. & Fritz, U. 2018. Phylogeography and taxonomy of the Barred Grass Snake (Natrix helvetica), with a discussion of the subspecies category in zoology. Vertebr. Zool. 68: 253–267.
- Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. 2018. MEGA x: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549.
- Leigh, J.W. & Bryant, D. 2015. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 6: 1110–1116.
- Lohman, D.J., Ingram, K.K., Prawiradilaga, D.M., Winker, K., Sheldon, F.H., Moyle, R.G., Ng, P.K.L., Ong, P.S., Wang, L.K., Braile, T.M. & Astuti, D. 2010. Cryptic genetic diversity in “widespread” Southeast Asian bird species suggests that Philippine avian endemism is gravely underestimated. Biol. Cons. 143: 1885–1890.
- Mattoccia, M., Romano, A. & Sbordoni, V. 2005. Mitochondrial DNA sequence analysis of the spectacled salamander, Salamandrina terdigitata (Urodela: Salamandridae), supports the existence of two distinct species. Zootaxa 995: 1–19.
- Myers, N., Mittermeier, R.A., Mittermeier, C.G., Da Fonseca, G.A. & Kent, J. 2000. Biodiversity hotspots for conservation priorities. Nature 403: 853–858.
- Neto, J.M., Arroyo, J.L., Bargain, B., Monros, J.S., Matrai, N., Prochazka, P. & Zehtindjiev, P. 2012. Phylogeography of a habitat specialist with high dispersal capability: the Savi’s Warbler Locustella luscinioides. PLoS One 7: e38497.
- Nieto, I., Chamorro, D., Palomo, L.J., Real, R. & Muñoz, A.-R. 2018. Is the Eurasian Reed Warbler Acrocephalus scirpaceus a regular wintering species in the Iberian Peninsula? Ringing data say yes. Acta Ornithol. 53: 61–68.
- Nissardi, S. 1998. Caso di svernamento di cannaiola, Acrocephalus scirpaceus, nello stagno di Molentargius (Sardegna meridionale). Riv. Ital. Ornitol. 68: 221–222.
- Nissardi, S. & Zucca, C. 2001. Complete post-breeding moult in Reed Warbler Acrocephalus scirpaceus in southern Sardinia. Ringing Migr. 20: 381–382.
- Olsson, U., Rguibi-Idrissi, H., Copete, J.L., Matos, J.L.A., Provost, P., Amezian, M., Alström, P. & Jiguet, F. 2016. Mitochondrial phylogeny of the Eurasian/African reed warbler complex (Acrocephalus, Aves). disagreement between morphological and molecular evidence and cryptic divergence: A case for resurrecting Calamoherpe ambigua Brehm 1857. Mol. Phylogenet. Evol 102: 30–44.
- Pavia, M., Galimberti, A., Pellegrino, I., Silvano, F., Zuccon, D. & Boano, G. 2018. New insights into the morphology and taxonomy of the Acrocephalus baeticatus / scirpaceus species complex based on a newly found West African syntopic population. Vie Milieu 68: 25–31.
- Pellegrino, I., Boatti, L., Cucco, M., Mignone, F., Kristensen, T.N., Mucci, N., Randi, E., Ruiz-Gonzalez, A. & Pertoldi, C. 2016. Development of SNP markers for population structure and phylogeography characterization in Little Owl (Athene noctua) using a genotyping-by-sequencing approach. Consev. Genet. Resour. 8: 13–16.
- Pellegrino, I., Cucco, M., Harvey, J.A., Liberatore, F., Pavia, M., Voelker, G. & Boano, G. 2017. So similar and yet so different: taxonomic status of Pallid Swift Apus pallidus and Common Swift Apus apus. Bird Study 64: 344–352.
- Pellegrino, I., Ilahiane, L., Boano, G., Cucco, M., Pavia, M., Prestridge, H.L. & Voelker, G. 2021. Avian haemosporidian diversity on Sardinia: a first general assessment for the insular Mediterranean. Diversity. (Basel) 13: 75.
- Pellegrino, I., Negri, A., Boano, G., Cucco, M., Kristensen, T.N., Pertoldi, C., Randi, E., Šálek, M. & Mucci, N. 2015. Evidence for strong genetic structure in European populations of the little owl Athene noctua. J. Avian Biol. 46: 462–475.
- Pertoldi, C., Pellegrino, I., Cucco, M., Mucci, N., Randi, E., Laursen, J.T., Sunde, P., Loeschcke, V. & Kristensen, T.N. 2012. Genetic consequences of population decline in the Danish population of the Little Owl (Athene noctua). Evol. Ecol Res. 14: 921–932.
- Pons, J.M., Cibois, A., Fournier, J., Fuchs, J., Olioso, G. & Thibault, J.C. 2019. Gene flow and genetic divergence among mainland and insular populations across the south-western range of the Eurasian treecreeper (Certhia familiaris, Aves). Biol. J. Linn. Soc. 126: 447–461.
- Pons, J.M., Thibault, J.C., Aymí, R., Grussu, M., Muntaner, J., Olioso, G., Sunyer, J.R., Touihri, M. & Fuchs, J. 2016. The role of western Mediterranean islands in the evolutionary diversification of the spotted flycatcher Muscicapa striata, a long-distance migratory passerine species. J. Avian Biol. 47: 386–398.
- Procházka, P., Stokke, B.G., Jensen, H., Fainová, D., Bellinvia, E., Fossøy, F., Vikan, J.R., Bryja, J. & Soler, M. 2011. Low genetic differentiation among reed warbler Acrocephalus scirpaceus populations across Europe. J. Avian Biol. 42: 103–113.
- Raković, M., Neto, J.M., Lopes, R.J., Koblik, E.A., Fadeev, I.V., Lohman, Y.V., Aghayan, S.A., Boano, G., Pavia, M., Perlman, Y., Kiat, Y., Dov, A.B., Martin Collison, J., Voelker, G. & Drovetski, S. 2019. Geographic patterns of mtDNA and Z-linked sequence variation in the common chiffchaff and the ‘chiffchaff complex’. PLoS One 14: e0210268.
- Scalercio, S., Cini, A., Menchetti, M., Vodă, R., Bonelli, S., Bordoni, A., Casacci, L.P., Dincă, V., Balletto, E., Vila, R. & Dapporto, L. 2020. How long is 3 km for a butterfly? Ecological constraints and functional traits explain high mitochondrial genetic diversity between Sicily and the Italian Peninsula. J. Anim. Ecol. 89: 2013–2026.
- Stewart, J.R., Lister, A.M., Barnes, I. & Dalén, L. 2010. Refugia revisited: individualistic responses of species in space and time. P. R. Soc. B-Biol. Sci. 277: 661–671.
- Stöck, M., Sicilia, A., Belfiore, N.M., Buckley, D., Brutto, S.L., Valvo, M.L. & Arculeo, M. 2008. Post-Messinian evolutionary relationships across the Sicilian channel: mitochondrial and nuclear markers link a new green toad from Sicily to African relatives. BMC Evol. Biol. 8: 56–74.
- Svensson, L. 1992. Identification Guide to European Passerines. 4th rev. British Trust for Ornithology, Stockholm.
- Villesen, P. 2007. Fabox: an online toolbox for fasta sequences. Mol. Ecol. Notes 7: 965–968.
- Voelker, G. & Light, J.E. 2011. Palaeoclimatic events, dispersal and migratory losses along the Afro-European axis as drivers of biogeographic distribution in Sylvia warblers. BMC Evol. Biol. 11: 163.
- Wauters, L.A., Amori, G., Aloise, G., Gippoliti, S., Agnelli, P., Galimberti, A., Casiraghi, M., Preatoni, D. & Martinoli, A. 2017. New endemic mammal species for Europe: Sciurus meridionalis (Rodentia, Sciuridae). Hystrix 28: 1–8.
- Zuccon, D., Pons, J.M., Boano, G., Chiozzi, G., Gamauf, A., Mengoni, C., Nespoli, D., Olioso, G., Pavia, M., Pellegrino, I., Raković, M., Randi, E., Idrissi, H.R., Touihri, M., Unsöld, M., Vitulano, S. & Brambilla, M. 2020. Type specimens matter: new insights on the systematics, taxonomy and nomenclature of the subalpine warbler (Sylvia cantillans) complex. Zool. J. Linn. Soc-Lond 190: 314–341.
