Abstract
To explore the composition, distribution and interrelation of the pineapple mealybugs in China, samples were collec from 14 counties of five provinces. The genetic variations of pineapple mealybugs from China, Brazil and Hawaii of USA have been analyzed. The whole ribosomal internal transcribed spacer 1 (ITS1), 5.8S and ribosomal internal transcribed spacer 2 (ITS2) were sequenced for all samples. Pink pineapple mealybug (PPM), Dysmicoccus brevipes (Cockerell) and gray pineapple mealybug (GPM), Dysmicoccus neobrevipes Beardsley are found causing damage on Ananas comosus (L.) Merr. PPM is a predominant species of mealybug on A. comosus in China. Though GPM has wider host range in China, PPM has larger distribution area than GPM. The results showed that two pineapple mealybug species belong to two different clades based on the sequences of ITS. PPM from China was found to have four haplotypes, and GPM had two haplotypes. The genetic variation of PPM is greater than that of GPM, though both of them had one predominant haplotype. One haplotype of PPM was found in samples from mainland China, and three from Hainan Island. Another haplotype of PPM was observed among the samples from Brazil and Hawaii. However, mealybugs from Wanning City of Hainan Island in China represented a different lineage that clearly diverged from other populations, which would be of a cryptic lineage or species in the pink pineapple mealybug complex. Mealybugs from Wanning City are probably native and were present long before the exotic pink pineapple mealybug was introduced in the early twentieth century. Most GPM, represented by a predominant haplotype, was widely found on sisal in plantations in south China. Other GPM, often found on sisal in urban green belt in south China, was of different haplotype from samples on sisal in plantations, probably originating from Taiwan.
Introduction
Pineapple mealybugs are cosmopolitan pests of Ananas comosus (L.) Merr and vectors of mealybug pineapple wilt disease which is a serious threat to commercial pineapple production. The host range of pink pineapple mealybug (PPM), Dysmicoccus brevipes (Cockerell) (Hemiptera: Pseudococcidae), an important pest of A. comosus and other bromeliads, is more than 50 plant families, including Annona squamosa L., Annona muricata L., Musa balbisiana Collawild, Apium graveolens L., Citrus spp., Coffea spp., Gossypium spp., Euphorbia spp., Gliricidia spp., Hibiscus spp., Paspalum conjugatum Bergius, Morus alba L., and Cyperu srotundus L. (Mau et al., 1992). Gray pineapple mealybug (GPM) was found by Ito (Citation1938), and named as Dysmicoccus neobrevipes Beardsley by Beardsley (Citation1959, Citation1965). PPM and GPM have similar external morphology. The ventral sclerotization of the anal lobes and the length of the setae on the dorsum of the abdomen are the primary characteristics used to differentiate the females of two species (Beardsley Citation1959). PPM and GPM differ not only in morphological characteristics, but also in reproduction and habitat. The male adult of PPM was found to be present in Madagascan and Brazil; however, the normal reproduction is parthenogenesis; meanwhile PPM normally feeds near the soil surface around the roots of the host plant, which is distinctly different from GPM (Beardsley Citation1959; Sether et al. Citation2005).
During the past century there has been concern about these two species because they have been implicated in transmitting the viral pineapple mealybug wilt disease (Rohrbach et al. Citation1988; Sether et al. Citation1998; Sether Citation2002). The presence of mealybug and closterovirus infection were found to be necessary for the development of mealybug wilt disease of A. comosus in Hawaii (Sether and Hu Citation2002; Sether et al. Citation2005). Pineapple mealybug wilt disease and pineapple mealybugs are commonly found on A. comosus in southern China, where GPM was widely known for being seriously damaging to Agave sisalana Perrine (Aspargales: Asparagaceae) in the last six years.
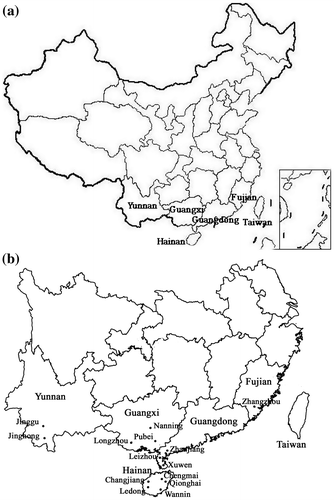
Sequence analysis of the rDNA ITS region is widely used in phylogeny and differentiation of populations, owing to the high degree of variation between populations or closely related species and the fact that it is easy to amplify from small quantities of DNA (Marcilla et al. Citation2001; Lantz et al. Citation2002; Xu et al. Citation2009; Kaura et al. Citation2010; Lindner and Banik Citation2011). Most mealybugs have large distribution area and wide host range, but little is known about the differentiation of mealybug populations. To determine genetic composition and relationship between PPM and GPM species, the ITS sequences of 16 pineapple mealybug populations from five provinces in China (Figure ) and one population from Hawaii were analyzed. This information will help to understand the population structure and molecular evolution of pineapple mealybugs, and prevent the introduction of other non-native genotypes into China (Saltonstall Citation2002).
Materials and methods
Processing of mealybugs
Pineapple mealybugs from A. comosus were obtained in 14 counties across five provinces in China, and a sample of PPM from Hawaii, USA was also included. In addition, Phenacoccus solenopsis Tinsley and Planococcus minor (Maskell) from China were used as outgroups in data analysis. Samples were collected in 2010–2012 (Table ).
Table 1. Collection data for samples of Dysmicoccus brevipes, Dysmicoccus neobrevipes and outgroup species information.
Live insects were carefully removed from the host plants and maintained without food for 24 h before storing in 95% ethanol at 4°C. Voucher samples have been preserved at the South Subtropical Crops Research Institute, Chinese Academy of Tropical Agricultural Sciences.
Samples were rinsed with double distilled water and then air dried. All specimens were examined under the microscope for the presence of parasitoids. Total DNA was extracted from single parasitoid-free adult females using the TIANamp Genomic DNA kit (Tiangen Biotech, Beijing, China).
Both 18S and 28S sequencing and morphological examination were conducted to differentiate between the two species of PPM and GPM. The 18S and 28s primers were obtained from Downie and Gullan (Citation2004) and Dietrich et al. (Citation2001) respectively. Accession numbers in GenBank for 18S sequences of the Wanning population and other PPM populations were JF965398, JF965399 respectively, and number for GPM is JF965400. Accession numbers in GenBank for 28S sequences of PPM and GPM were JF965409, JF965410 respectively. The morphological examination was done by Prof. Sanan Wu, Beijing Forestry University.
Template preparation and DNA manipulation
The primers of ITS F: 5′-CGTTGATTACGTCCCTGCCCTTTG-3′ and ITS R: 5′-TGCTTAAGTTCAGCGGGTAG-3′ were obtained from GenBank (EU307273, Chiu et al. Citation2001; GU134673, Malausa et al. Citation2010). PCR amplification was performed in 25 μl reaction volumes with 3 μl of template DNA. The PCR reaction mix contained 2.5 μl 10× Taq reaction buffer (Promega, Madison, WI, USA), 0.5 U Taq DNA polymerase (Promega), 3.5 mM MgCl2, and 15 pM of each primer. PCR reaction conditions were as follows: 95°C for 3 min, followed by 34 cycles of 95°C for 30 s, 55°C for 45 s, 72°C for 1.5 min, and a final extension of 5 min at 72°C.
Five microliters of each PCR product were run on a 1% agarose gel to determine the presence and size of amplified DNA. PCR products were sequenced in both forward and reverse directions, and to achieve the whole ITS sequences, new primers need to be designed from the intermediate sequence. Amplification products were purified and sequenced by Invitrogen Biotechnology (Shanghai, China) on both strands using PCR primers. Sequences of all haplotypes of PPM and GPM have been deposited in GenBank under accession numbers JX228127–JX228133; accession numbers of Planococcus minor and Phenacoccus solenopsis were JX228134 and JX228135 respectively. The internal transcribed spacer 2 gene sequences of PPM from Brazil were obtained from GenBank with accession number GU134673, and the internal transcribed spacer 1 homologous sequence of PPM from Taiwan was obtained from GenBank with accession number EU307273.
Data analysis
Sequences were aligned using ClustalX (version 2.0) and unique haplotypes were identified with Arlequin (version 3.5). Descriptive statistics (number of variable sites, number of haplotypes, haplotype diversity, nucleotide diversity, average number of nucleotide differences between haplotypes) were calculated using DNASP (version 5.0), and Fst were calculated using Arlequin version 3.5 (Excoffier et al. Citation2005).
The genealogical relationships among pink pineapple mealybugs ITS2 haplotypes were studied by constructing a network based on the statistical parsimony method of Templeton et al. (Citation1992), using the software TCS 1.20 (Clement et al. Citation2000). A network can be a more appropriate way of depicting intraspecific gene genealogies than a bifurcating tree because of the potential for extinct ancestral nodes and multifurcating relationships (Posada and Crandall Citation2001).
Phylogenetic analyses were performed by maximum parsimony (MP) analysis and neighbor joining (NJ) analysis by using PAUP* 4.10 (Swofford Citation2003). Incongruence among data partitions was analyzed using a partition homogeneity test with 1000 replications as implemented in PAUP*. MP and NJ analyses used the heuristic search option with tree-bisection-reconnection (TBR) branch swapping, collapsing zero-length branches, and equal weighting of all characters. MP bootstrap support was calculated using 1000 replicates.
Results
The occurrence of two species of pineapple mealybugs in China
The pink pineapple mealybug (PPM) is a predominant species of mealybugs on A. comosus in China, occurring throughout the pineapple planting areas. PPM is normally found on the base of the leaf or stem or on the roots of pineapple plants, and is also found feeding at the base of crowns or on the dents of pineapple fruits.
The gray pineapple mealybug (GPM) occurs in Hainan and Guangdong province. The host plants include Agave sisalana, Agave americana, Ananas comosus, and Musa nana, however, only one pineapple orchard at Ledong in Hainan Island was found to be damaged by GPM. GPM is generally found on the aerial portions of the host plants, including the leaves of Agave spp., the fruits or fruit stalks of A. comosus and M. acuminata, but not on the roots.
The genetic structure of two species of pineapple mealybugs
According to ITS1 and ITS2 sequences, the populations of PPM from mainland China have four haplotypes, which are QJ, QH, WN1 and WN2. The population from Wanning (Hainan, China) has two closely related haplotypes, which are WN1 and WN2 (Table ). Most pink pineapple mealybug populations in China are classified to the haplotype QJ, samples showing the haplotype QH, WN1 and WN2 are mostly from the Hainan Island (Table ). Another haplotype was observed among the sample from Hawaii, the same ITS2 sequences were found from PPM samples in Hawaii and Brazil (GU134673).
Table 2. Distribution frequency of different haplotypes in geographical populations of Dysmicoccus brevipes and Dysmicoccus neobrevipes.
As for GPM from mainland China, only two haplotypes based on ITS1 and ITS2 were found, which are JM and XS. Most gray pineapple mealybug populations in China are grouped to the haplotype JM, which is the most widely distributed. The genetic diversity indexes in Table showed that PPM has more complex genetic diversity than that of GPM. The interesting fact is that the ITS1 sequence of PPM in Taiwan (EU307273) is the same as the sequence of GPM in Xiashan, Zhanjiang, Guangdong, with little variation from other populations of GPM in China (Figure ).
Table 3. Genetic diversity indexes of Dysmicoccus brevipes and Dysmicoccus neobrevipes.
The genetic differentiation of two species of pineapple mealybugs
Most populations of PPM and GPM have no any genetic differentiation. According to the FST values of pairwise populations of two species of pineapple mealybugs, only Wanning and Hawaii populations of PPM and Zhanjiang population of GPM showed a high genetic differentiation (Tables , ). Genetic distances between species were obviously greater than intraspecific genetic distances (Table ).
Table 4. FST values of pairwise populations of Dysmicoccus brevipes.
Table 5. FST values of pairwise populations of Dysmicoccus neobrevipes.
Table 6. Genetic distances among different haplotypes of Dysmicoccus brevipes and Dysmicoccus neobrevipes.
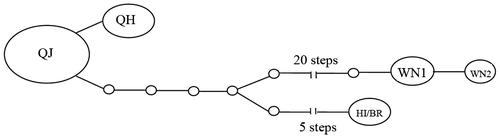
It is indicated by the TCS haplotype network that haplotype QJ, found in most mealybug samples from mainland China, is closely related to haplotype QH. The haplotype network also revealed a very high divergence of the haplotype WN1 and WN2 compared to all other haplotypes of PPM (Figure ). The genetic distances in Table also illustrated that population of PPM from Wanning has most distant genetic relationship with other populations of PPM.
Figure 2. TCS haplotype network showing genealogical relationships among 12 populations of Dysmicoccus brevipes. Haplotypes are connected with a 90% CL. White dots represent mutational steps separating the observed haplotypes, and the size of each oval is proportional to the frequency of the haplotypes in the analysis.
Two species of pineapple mealybugs obviously belong to two different clades based on the ITS sequences. EU307273 is the homology sequence of PPM from Taiwan, but it unexpectedly belongs to the clade of GPM (Figure ). PPMs in China are clustered into three different major clades; most samples of QJ clade are from mainland China, most samples of QH clade are from Qionghai, Hainan Island, and most samples of WN clade are from Wanning, Hainan Island. GPM in China is clustered into two different major clades; most samples of JM clade are from Hainan Island and Leizhou, Guangdong province, and most samples of XJ clade are from Zhanjiang, Guangdong province (Figures , ).
Figure 3. Phylogenetic trees of Dysmicoccus brevipes and Dysmicoccus neobrevipes based on ITS1 sequences. On the left is the MP tree, and on the right is the NJ tree. PAUP 4.10 software was used for the analysis. Planococcus minor and Phenacoccus solenopsis were included as outgroups. EU307273 is the homology sequence of pink pineapple mealybug from Taiwan.
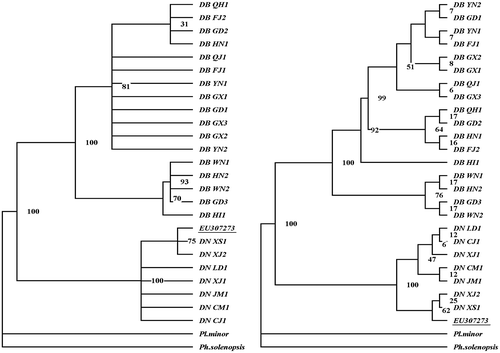
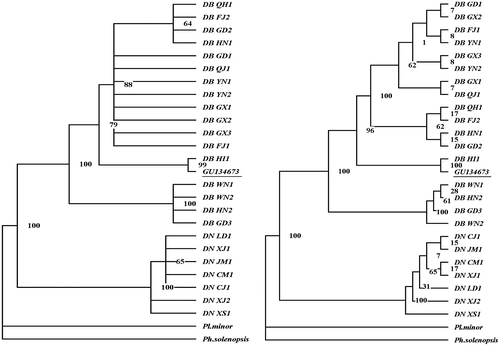
Figure 4. Phylogenetic trees of Dysmicoccus brevipes and Dysmicoccus neobrevipes based on ITS2 sequences. On the left is the MP tree, and on the right is the NJ tree. PAUP 4.10 software was used for the analysis. Planococcus minor and Phenacoccus solenopsis were included as outgroups. GU134673 is the homology sequence of pink pineapple mealybug from Brazil.
Discussion
More than 50 families host plants can be damaged by pink pineapple mealybug (PPM) (Mau and Martin Kessing Citation1992). Arachis hypogaea L. was reported as being damaged by PPM in Taiwan (Huang et al. Citation2002), but A. comosus is currently the only host plant found with damage caused by PPM in mainland China. Gray pineapple mealybug (GPM) has a short history of damaging A. comosus in individual orchards in mainland China, but has been a serious threat to commercial sisal production because it results in a devastating disease, whose symptoms are like mealybug wilt of pineapple. PPM has a longer history in China, with larger distribution area and higher genetic differentiation than GPM. A habitual mistake has been to misidentify GPM as PPM on pineapple, because of their similar morphology. PPM from Taiwan has the same ITS1 sequence (accession number: EU307273) as GPM from Zhanjiang, Guangdong province.
The relationships among ITS lineages of PPM populations from mainland China reveal their biogeographic patterns. Four haplotypes were found in China: one from mainland China and three from the island of Hainan. Pink pineapple mealybugs collected in Yunnan province, Guangxi Zhuang Autonomous Region, and most samples from Guangdong, Fujian province, shared an identical haplotype. Two haplotypes of GPM were found in China, and the most common haplotype, DN_JM, was observed in 71% of the population samples. GPM presented by haplotype DN_XS shares the same sequence with EU307273, which probably originated in Taiwan, if sequence of EU307273 was actually of GPM and not PPM in Taiwan. This all suggests that limited genetic variation existed in pineapple mealybugs in mainland China.
ITS sequences of PPM from Wanning are significantly different from other populations. However, morphological examination failed to prove that PPM from Wanning is a different species. PPM from Wanning shares the same fragment of 28S with other populations; the fragment of 18S is only slightly different with one base transition at the 271st base. The genetic distance between the collections from Wanning and those from other populations suggest that PPM may be a species complex which includes at least two cryptic lineages or sibling species. The PPM from Wanning is of a cryptic lineage or species in the complex.
Phylogenetic analysis suggested that the PPM from Hawaii was a lineage of mealybugs living predominantly in Brazil. The mealybugs sharing an identical haplotype in mainland China were most closely related to the mealybug population from Qionghai. There would be a more recent common ancestor of most samples of PPM from mainland China with samples of PPM from Hawaii, but not with samples of PPM from Wanning. The analysis based on mitochondrial cytochrome oxidase I (COI) indicated that PPM from Hawaii was very likely of the mealybug lineage from Thailand. The mealybugs sharing an identical haplotype with PPM from mainland China were most closely related to the mealybug population from the Philippines, followed by Hawaii and Thailand (He et al. Citation2012). All of these suggest that most PPM in mainland China are probably introduced by human activities from Southeast Asia, and PPM in Southeast Asia might have originate from Central and South America. It also can be contended that the lineage present in Wanning is native or originated from other countries or regions not mentioned above. More sources of PPM throughout the world should be studied in order to understand the invasion history of pink pineapple mealybug into China and the migrating pathway of pink pineapple mealybug around the world.
The internal transcribed spacer (ITS) was widely used in determination of species. By revealing hidden diversity, it has proven especially powerful in identifying fungi. However, it has limited use for environmental barcoding due to paralogs, the potential for unidentifiable chimaeras and priming across taxa (Schoch et al. Citation2012; Stern et al. Citation2012). Base deletion of ITS sequences is common in analyses of population structure, molecular evolution and phylogenetics, which commonly lead to a fragment length difference. For Anopheles subpictus Grassi, ITS2 sequence in Chandigarh was 681 bp compared to 491 bp in Hoshiarpur, Punjab, India (Kaura et al. Citation2010). The result based on ITS sequences of pineapple mealybugs in this study is consistent to previous research for COI sequences. We found two haplotypes based on ITS sequences from the Wanning population of pineapple mealybugs in this study, instead of one haplotype based on COI sequences, suggesting that not only COI but also ITS can be used to study the genetic differentiation.
The recognition of several distinct lineages indicates a need to identify and prevent the introduction of genotypes to a new region (Havill et al. Citation2006). Pink pineapple mealybug was found from A. comosus at Kingston, Jamaica prior to 1893. It is believed to have the same origin as A. comosus, i.e. South America or the New World (Ferris Citation1950; Sether Citation2002). To determine if there are distinct and distant populations of wild pineapple from Wanning, Hainan, as for the pink pineapple mealybug, further studies are required.
Acknowledgments
This work was supported in part by Special Fund for Agri-Scientific Research in the Public Interest (201203021), Natural Science Foundation of Hainan Province of China (313053) and The Youth Foundation of National Natural Science Foundation of China (31300348). We are grateful to Dr. WF Zhao, Prof. YQ Liu & JX Zheng, for helping to collect the insect samples. Special thanks to Dr. Diane M. Sether and Prof. John S. Hu, for providing mealybug samples of Hawaii. The authors are also thankful to Prof. Y Chen and two anonymous reviewers for their valuable comments in improving the quality of the present research work.
References
- Beardsley JW. 1959. On the taxonomy of pineapple mealybugs in Hawaii, with a description of a previously unnamed species (Homoptera: Pseudococcidae). Proc Hawaii Entomol Soc. 17(1):29–37.
- Beardsley JW. 1965. Notes on the Pineapple mealybug complex, with descriptions of two new species (Homoptera: Pseudococcidae). Proceddings of the Hawaiian. Entomological Society. 19(1):55–68.
- Chiu YC, Wu WJ, Shi CJ. 2001. Identification of three Aulacaspis species (Homoptera: Diaspididae) by PCR-RFLP analysis for quarantine application. Formos Entomol. 21(4):365–375 ( In Chinese).
- Clement M, Posada D, Crandall KA. 2000. TCS: a computer program to estimate gene genealogies. Mol Ecol. 9(10):1657–1660.
- Cockerell TDA. 1893. The West Indian species of Dactylopius. The Entomologist. 26:177–179.
- Dietrich CH, Rakitov RA, Holmes JL, Black IVWC. 2001. Phylogeny of the major lineages of Membracoidea based on 28S rDNA sequences. Mol Phylogenet Evol. 18(2):293–305.
- Downie DA, Gullan PJ. 2004. Phylogenetic analysis of mealybugs (Hemiptera: Coccoidea: Pseudococcidae) based on DNA sequences from three nuclear genes, and a review of the higher classification. Syst Entomol. 29(2):238–260.
- Excoffier Laval LG, Schneider S. 2005. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinform Online. 1:47–50.
- Ferris GF. 1950. Atlas of the scale insects of North America. Stanford, CA: Stanford University Press; p. 59–60.
- Havill NP, Montgomery ME, Yu GY, Shiyake S, Caccone A. 2006. Mitochondrial DNA from hemlock woolly adelgid (Hemiptera: Adelgidae) suggests cryptic speciation and pinpoints the source of the introduction to Eastern North America. Ann Entomol Soc Am. 99(2):195–203.
- He YB, Wan XW, Liu YH, Sun GM, Zhan RL. 2012. Mitochondrial COI from Dysmicoccus brevipes (Hemiptera: Pseudococcidae) suggests cryptic lineage and pinpoints the source of the introduction to China. Fla Entomol. 95(1):183–191.
- Huang SH, Wong CY, Cheng CH. 2002. A newly recorded insect pest, pink pineapple mealybug (Dysmicoccus brevipes (Cockerell)) (Hemiptera: Pseudococcidae), infecting on the roots of peanut in Taiwan. Plant Protect Bull. 44(2):141–146.
- Ito K. 1938. Studies on the life histories of the pineapple mealybug, Pseudococcus brevipes (Ckll.). J Econ Entomol. 31(2):291–298.
- Kaura T, Sharma M, Chaudhry S, Chaudhry A. 2010. Sequence polymorphism in spacer ITS2 of Anopheles (Cellia) subpictus Grassi (Diptera: Culicidae). Caryologia. 63(2):124–133.
- Lantz H, Andreasen K, Bremer B. 2002. Nuclear rDNA ITS sequence data used to construct the first phylogeny of Vanguerieae (Rubiaceae). Plant Syst. Evol. 230(3–4):173–187.
- Larkin MA, Blackshields G, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25(11):1451–1452.
- Lindner DL, Banik MT. 2011. Intragenomic variation in the ITS rDNA region obscures phylogenetic relationships and inflates estimates of operational taxonomic units in genus Laetiporus. Mycologia. 103(4):731–740.
- Malausa T, Fenis A, Warot S, Germain JF, Ris N, Prado E, Botton M, Vanlerberghe-Masutti F, Sforza R, Cruaud C, Couloux A, Kreiter P. 2010. DNA markers to disentangle complexes of cryptic taxa in mealybugs (Hemiptera: Pseudococcidae). J. Applied Entomol. 135(1–2):142–155.
- Marcilla A, Bargues MD, Ramsey JM, Magallon-Gastelum E, Salazar-Schettino PM, Abad-Franch F, Dujardin JP, Schofield CJ, Mas-Coma S. 2001. The ITS-2 of the nuclear rDNA as a molecular marker for populations, species, and phylogenetic relationships in Triatominae (Hemiptera: Reduviidae), vectors of Chagas disease. Mol Phylogenet Evol. 18(1):136–142.
- Mau RFL, Martin Kessing JL. 1992. Dysmicoccus brevipes (Cockerell). http://www.extento.hawaii.edu/kbase/crop/type/d_brevip.htm
- Posada D, Crandall KA. 2001. Intraspecifc gene genealogies: trees grafting into networks. Trends Ecol Evol. 16(1):37–45.
- Qin ZQ, Wu JH, Ren SX, Wan FH. 2010. Risk analysis of the alien invasive gray pineapple mealybug (Dysmicoccus neobrevipes Beardsley) in China. Scientia Agricultura Sinica. 2010,43(3):626–633 ( In Chinese).
- Rohrbach KG, Beardsley JW, German TL, Reimer N, Sanford WG. 1988. Mealybug wilt, mealybugs, and ants on pineapple. Plant Dis. 72(7):558–565.
- Saltonstall K. 2002. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proc Natl Acad Sci USA. 99(4):2445–2449.
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque A, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. PNAS. 109(16):6241–6246.
- Sether DM. 2002. Pineapple mealybug wilt associated viruses: vectors, impacts, and dynamics [doctoral dissertation]. Honolulu, HI: University of Hawaii.
- Sether DM, Hu JS. 2002. Closterovirus infection and mealybug exposure are necessary for the development of mealybug wilt of pineapple disease. Phytopathology. 92(9):928–936.
- Sether DM, Melzer MJ, Busto J, Zee F, Hu JS. 2005. Diversity and mealybug transmissibility of ampeloviruses in pineapple. Plant Dis. 89(5):450–457.
- Sether DM, Ullman DE, Hu JS. 1998. Transmission of pineapple mealybug wilt-associated virus by two species of mealybugs (Dysmicoccus spp.). Phytopathology. 88(11):1224–1230.
- Stern RF, Andersen RA, Jameson I, Küpper FC, Coffroth M-A, Vaulot D, Gall FL, Véron B, Brand JJ, Skelton H, Kasai F, Lilly E, Keeling PJ. 2012. Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker. PLoS ONE. 7(8):e42780. doi:10.1371/journal.pone.0042780.
- Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer.
- Templeton AR, Crandall KA, Sing CF. 1992. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 132(10):619–633.
- Xu H, Wang ZT, Cheng KT, Wu T, Gu LH, Hu ZB. 2009. Comparison of rDNA ITS sequences and tanshinones between Salvia miltiorrhiza populations and Salvia species. Bot Stud. 50(2):127–135.