 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The construction of decision-theoretical Bayesian designs for realistically complex nonlinear models is computationally challenging, as it requires the optimization of analytically intractable expected utility functions over high-dimensional design spaces. We provide the most general solution to date for this problem through a novel approximate coordinate exchange algorithm. This methodology uses a Gaussian process emulator to approximate the expected utility as a function of a single design coordinate in a series of conditional optimization steps. It has flexibility to address problems for any choice of utility function and for a wide range of statistical models with different numbers of variables, numbers of runs and randomization restrictions. In contrast to existing approaches to Bayesian design, the method can find multi-variable designs in large numbers of runs without resorting to asymptotic approximations to the posterior distribution or expected utility. The methodology is demonstrated on a variety of challenging examples of practical importance, including design for pharmacokinetic models and design for mixed models with discrete data. For many of these models, Bayesian designs are not currently available. Comparisons are made to results from the literature, and to designs obtained from asymptotic approximations. Supplementary materials for this article are available online.
1. Introduction
Bayesian design of experiments is a natural paradigm for many problems arising in the physical sciences and engineering, particularly those concerning the estimation of nonlinear models where design performance, as measured by classical optimality criteria, is dependent on the a priori unknown values of the model parameters. A decision-theoretic approach, reviewed by Chaloner and Verdinelli (Citation1995), determines an optimal allocation of experimental resources via maximization of the expected utility
(1)
(1) Here, the utility u(δ, ψ, y) quantifies the experimenter's gain from using design
to obtain data
assuming model parameter values ψ ∈ Ψ, with the statistical model defined through the joint density function π(y, ψ | δ) = π(y | ψ, δ)π(ψ). As an example, assume the ith response is modeled as yi = g(xi; θ) + ϵi with the xi defining values taken by a controllable variable, θ being a vector of unknown parameters defining the mean response, and observation error ϵi ∼ N(0, σ2) (i = 1, …, n). Then ψT = (θT, σ2), δ = (x1, …, xn)T, and likelihood π(y | ψ, δ) is a multivariate normal density function. The utility function u(δ, ψ, y) will typically be a function of some posterior quantities of ψ (see Section 3.1).
Selection of a fully Bayesian optimal design has traditionally been challenging for all but the most straightforward utility functions and models due to the high-dimensional and, typically, analytically intractable integrals in (Equation1
(1)
(1) ). Some recent progress has been made using simulation-based methodologies for low-dimensional problems, that is, small numbers of controllable variables and/or small numbers of design points, see Ryan et al. (Citation2016) and references therein. There are, however, no methods available for decision-theoretic Bayesian optimal, or near-optimal, multi-variable design for nonlinear models. The methodology in this article fills this important gap, and is demonstrated on generic problems of practical importance including pharmacokinetic studies and experiments that produce discrete data. Previous attempts to obtain fully Bayesian optimal designs for these types of experiment have been extremely limited.
In a landmark article for low-dimensional design problems, Müller and Parmigiani (Citation1996) proposed selection of a design by maximizing a surrogate function found by approximating U(δ) for a small number, m, of designs using simulation, and then smoothing the resulting values, ). See also Jones et al. (Citation2016) and Weaver et al. (Citation2016). In essence, these approaches perform a computer experiment to construct a statistical emulator for the approximation
, a research area where there has been huge activity in recent years (see, e.g., Dean et al. Citation2015, sec. V). For an experiment with n runs and v variables, δ has nv elements. Therefore, application of this approach to design for multi-variable models suffers from a curse of dimensionality, requiring (i) the construction of emulators in very high dimensions; (ii) large, for example, space-filling, designs composed of selections of points from an nv-dimensional space, leading to (iii) a prohibitive number of evaluations of
, particularly if
is computationally expensive.
Our approach overcomes these problems by building a series of one-dimensional emulators for the approximated expected utility. We emulate as a function of only the ith “coordinate” (or element) δi conditional on δ(i) = (δ1, …, δi − 1, δi + 1, δnv)T, the values of all coordinates excluding the ith (i = 1, …, nv). When these emulators are combined with a continuous version of the coordinate exchange algorithm (Meyer and Nachtsheim Citation1995), an effective and computationally efficient design selection methodology results. Conditional, coordinate-wise, optimization is key to overcoming the curse of dimensionality described above.
Until relatively recently, the usual approach to Bayesian design was to use a normal distribution as an asymptotic approximation to the posterior distribution of ψ (e.g., Chaloner and Larntz Citation1989). For standard utility functions (see Section 3.1.2), use of such a pseudo-Bayesian approach leads to the integrand in (Equation1(1)
(1) ) no longer depending on the data y. The resulting integral, with respect to ψ, typically has much lower dimension and can be approximated using efficient deterministic quadrature rules (Gotwalt, Jones, and Steinberg Citation2009). However, the appropriateness of such approximations for small experiments is open to question.
For high-dimensional design, an alternative to the use of a normal approximation was suggested by Ryan et al. (Citation2014). These authors combined the simulation-based approach of Müller (Citation1999) (see also Müller, Sanso, and De Iorio Citation2004; Amzal et al. Citation2006) with a dimension-reduction scheme to find designs for single-variable nonlinear models (v = 1) variables and a large number of runs. Designs were restricted to those formed from a sampling scheme defined via two parameters, for example, the initial design point and a spacing parameter. An optimal design in this subclass then consists of the best choices of these two parameters, a substantially easier optimization problem to solve.
In contrast to either applying an asymptotic approximation or restricting attention to a subset of the design space, both of which may result in the selection of inefficient designs with respect to the exact expected utility, we attempt to find optimal or efficient designs for the original problem across the whole design space via an approximate optimization scheme. These three different approaches are compared in Section 3.
The remainder of the article is organized as follows. In Section 2, we describe approximate coordinate exchange for finding decision-theoretic Bayesian designs, including the use of Monte Carlo integration and Gaussian process emulators to approximate the expected utility. The methods are applied to a range of challenging and practically relevant examples in Section 3 including models for which Bayesian design has previously been computationally infeasible. We summarize the advantages of our approach in Section 4 and highlight some ongoing work.
2. Approximate Coordinate Exchange (ACE)
We first establish some notation. Suppose that a design consists of n runs or points, each of which determines the settings of v controllable variables and results in a single observation of the response variable. Let D denote the n × v design matrix with kth row dk specifying the settings of the v factors in the kth run (k = 1, …, n). Let q = nv, then the design may be represented as a q-vector , where vec( · ) denotes vectorization via stacking the columns of a matrix and
is the q-dimensional design space.
The proposed algorithm for decision-theoretic Bayesian design has two phases. Phase I applies a novel coordinate exchange algorithm where, for each coordinate, maximization of U(δ) is replaced by maximization of a surrogate function . Phase I tends to produce clusters of design points that are very similar in the values of the controllable variables. Such clustering is common in heuristic design search (see also Gotwalt, Jones, and Steinberg Citation2009). Hence, in Phase II, we check if the points in each cluster can be consolidated into a replicated design point using a point exchange algorithm (Atkinson, Donev, and Tobias Citation2007, chap. 12). Replication of points is common in optimal design for parametric models and a key principle of design of experiments (Wu and Hamada Citation2009, chap. 1). In Phase II, the candidate set is the design found from Phase I. The two phases form an approximate coordinate and point exchange algorithm which, for brevity, we call the ACE algorithm.
In Section 2.1, we define the ACE algorithm. For Steps 2a–2c of the algorithm, we assume the availability of (i) a Monte Carlo approximation of the expected utility,
(2)
(2) with {yl, ψl}Bl = 1 a random sample from the joint distribution with density π(y, ψ | δ); (ii) coordinate-designs
at which we evaluate
, where
is the domain for the ith coordinate; and (iii) a suitable one-dimensional emulator,
, for
. Further details are given in Section 2.2, with examples in Section 2.4.
ACE is designed to solve a stochastic optimization problem, as only approximations to the expected utility are available formed as linear combinations of realizations of the random variable u(δ, y, ψ). As such, proposed changes to the design in Steps 2d and 4e of the algorithm are accepted with probability derived from a Bayesian test of the difference in the means of Monte Carlo approximations to the expected utility for the current and proposed designs. Further details are given in Section 2.3.
The R package acebayes (Overstall et al. Citation2017) provides an implementation of ACE and is available on CRAN.
2.1 The ACE Algorithm
| 1. | Choose an initial design δ0 and set the current design δC = δ0.
| ||||||||||||||||||||||||||||||||||
| 2. | For i = 1, …, q,
| ||||||||||||||||||||||||||||||||||
| 3. | Repeat Step 2 NI times.
| ||||||||||||||||||||||||||||||||||
| 4. |
| ||||||||||||||||||||||||||||||||||
| 5. | Repeat Step 4 NII times. | ||||||||||||||||||||||||||||||||||
| 6. | Return δ⋆ = δC. | ||||||||||||||||||||||||||||||||||
The decision on when to terminate a run of the algorithm, that is, choice of NI and NII, is complicated by the stochastic nature of the approximation to the expected utility. For the examples in Section 3, NI = 20 and NII = 100 are sufficient to achieve approximate convergence. Here, convergence is assessed graphically from trace plots of against iteration number; see Section 3.2 for examples of such plots.
To avoid local optima, the algorithm is run M times (in embarrassingly parallel fashion) with each run starting from a different, randomly chosen, initial design δ0 (a random Latin hypercube design, unless otherwise stated). The selected design, δ⋆, is the design having the highest average approximate expected utility, averaged across C sets of Monte Carlo simulations. In this article, M = C = 20 was used, unless otherwise stated.
2.2 Emulation via Computer Experiments (Steps 2a–2c)
In Phase I of the algorithm, a sequence of one-dimensional emulators is constructed for , i = 1, …, q (Step 2c). A variety of smoothing or interpolation techniques could be applied to construct each emulator. Müller and Parmigiani (Citation1996) used local polynomial regression to emulate low-dimensional design utilities. We adopt a Gaussian Process (GP) regression model (see, e.g., Rasmussen and Williams Citation2006), which is widely used for computer experiments, and use the posterior predictive mean as an emulator. Let
and
for any
. The GP model assumes that any vector
, for
and integer m0, has joint distribution
, with
the m0 zero-vector and A(ζ) an m0 × m0 covariance matrix. Hence, the posterior predictive mean of
at an arbitrary
can be derived using standard results on the conditional distribution of normal random variables and used as an emulator:
Under the common assumption of a squared exponential correlation function, A(ξi) and a(δ, ξi) have entries
for s, t = 1, …, m, where I(E) is the indicator function for the event E, and ρ, η > 0 are unknown parameters. The inclusion of nugget η ensures the emulator will smooth, rather than interpolate, the Monte Carlo approximations of the expected utility. To limit computational complexity, at each iteration we find maximum likelihood estimates of ρ and η via Fisher scoring (see, e.g., Pawitan Citation2001, pp. 174–177). In contrast, a fully Bayesian approach would require application of a Markov chain Monte Carlo algorithm to construct each emulator, substantially increasing the computational cost of the algorithm.
At each iteration of Step 2a, a coordinate-design ξi = (δ1i, …, δmi) must be chosen at which to evaluate . We use a space-filling design, specifically a randomly selected one-dimensional Latin hypercube design (see, e.g., Santner, Williams, and Notz Citation2003, chap. 5), constructed by dividing
into m equally sized sub-intervals, and then generating a point at random from each interval. We set m = 20, unless otherwise stated. This choice of m is conservative relative to the rule of thumb (Loeppky, Sacks, and Welch Citation2009) of setting m equal to 10 times the number of input dimensions (suggesting m = 10 in our case). We have, however, found it works well in practice for a variety of different types of examples, giving accurate emulators and not being overly computationally demanding.
2.3 Adjusting a Design Coordinate (Step 2d) or Point (Step 4e)
To make a change to the ith coordinate in Step 2d, we first find δ†i, the value of the coordinate that maximizes the emulator. We find the maximum by evaluating for 10,000 uniformly generated points in
. This discretization of the problem has proved both more reliable than continuous optimization and sufficiently computationally efficient.
Choice of δ†i is subject to both Monte Carlo error, from the evaluation of , and emulator error from the estimation of
resulting, for example, from an inappropriate choice of correlation function or errors in estimating ρ and η. It is clearly impossible to use usual residual diagnostics (Bastos and O’Hagan Citation2009) to check emulator adequacy at each iteration of the algorithm. Instead, emulator error is eliminated from the decision to adjust a design coordinate by performing additional Monte Carlo integration to calculate the probability p†I in (Equation3
(3)
(3) ). This quantity is the posterior probability that
under noninformative prior distributions and using Monte Carlo samples {ψCl, ylC}Bl = 1 and {ψ†l, y†l}Bl = 1, assuming both u(ψ†, y†, δC†) and u(ψC, yC, δC) are normally distributed with equal variances. See also Wang and Zhang (Citation2006) for use of a classical hypothesis test in a simulated annealing algorithm. If this normality assumption were severely violated, a more sophisticated test procedure could be adopted at greater computational cost.
A similar test is performed at Step 4e in Phase 2 of the algorithm to calculate p†II in (Equation4(4)
(4) ). We demonstrate the effect of Step 2d in the next section.
2.4 Illustrative Example
In this section, we illustrate the ACE methodology, in particular the combination of Steps 2c and 2d in selecting and accepting a proposed change to the design. To enable assessment of the algorithm, we consider the analytically tractable problem of finding a one-point optimal design for the single-variable Poisson model y|β ∼ Poisson(eβx). There is a single design coordinate, δ = x ∈ [ − 1, 1], and hence our notation is simplified by replacing δ by x in this example. A priori, we assume β ∼ N(0.5, 1) and adopt the utility function that leads to pseudo-Bayesian D-optimality (Section 3.1.2), given by
where
denotes the Fisher information. The expected utility is U(x) = 2log |x| + 0.5x and the optimal design is x⋆ = 1.
To simulate one iteration of Phase I of the ACE algorithm, we generate a coordinate-design ξ1 = {x1, …, xm} as a Latin hypercube and, for each xj, evaluate
where {βl}Bl = 1 for B = 2 is a sample from an N(0.5, 1) distribution. shows U(x) plotted against x with the points
and the GP emulator
superimposed (Steps 2a, 2b, and 2c). Clearly
is maximized at x† = 1, and hence this candidate point should be compared to the current point xC (Step 2d). shows the median posterior probability, p†I, of accepting this candidate point against xC, calculated from repeated calculation of (Equation3
(3)
(3) ) for multiple Monte Carlo samples. This probability is very close to one for nearly all values of xC except for xC ≈ x†, where the probability reduces to 1/2.
Figure 1. Poisson example in Section 2.4. (a), (c) expected utility U(x) against x, with Monte Carlo evaluations at the coordinate-design points and GP emulator
; (b), (d) median probability p†I of accepting the candidate point against the current point, xC. [Coordinate-designs are: ξ1 for (a), (b); ξ2 for (c), (d)].
![Figure 1. Poisson example in Section 2.4. (a), (c) expected utility U(x) against x, with Monte Carlo evaluations U˜(x) at the coordinate-design points and GP emulator U^(x); (b), (d) median probability p†I of accepting the candidate point against the current point, xC. [Coordinate-designs are: ξ1 for (a), (b); ξ2 for (c), (d)].](/cms/asset/c8e39f62-1517-4820-8ba3-e5088fa2014f/utch_a_1251495_f0001_b.gif)
For a second coordinate-design, ξ2 (a different Latin hypercube), the results in and are obtained. Here, the GP emulator could be viewed as inadequate with the estimate of η being too small, resulting in near interpolation of the . From ,
is maximized at x† = −1 and hence this becomes the candidate point. The median posterior acceptance probability, , is now only close to one if
is low, that is, |xC| < 0.5. Crucially, x† will be rejected with high probability if xC is close to the optimal design; at xC = 1, the probability drops to zero.
3. Substantive Examples
The ACE algorithm is now used to find decision-theoretic Bayesian designs for three important cases: a compartmental model, (hierarchical) logistic regression, and dose–response under model-averaging. The designs are found for commonly used utility functions and, where possible, compared to existing results.
The supplementary material for this article provides a detailed vignette that demonstrates the use of the acebayes package to find designs for the following examples, and R code to allow their straightforward reproduction.
3.1 Utility Functions
In this section, we assess and compare designs found using variants on two utility functions, Shannon information gain (SIG) and negative squared error loss (NSEL). In practice, the form of the chosen utility function should be driven by the aims of the experiment and may often incorporate a cost function. We assume throughout that the model parameters can be expressed as ψT = (θT, γT), with θ a p-vector of parameters of interest and γ a (P − p)-vector of nuisance parameters.
The SIG utility for θ is given by
(5)
(5) where (Equation5
(5)
(5) ) follows from an application of Bayes’ theorem and is often more useful for computation. A SIG-optimal design maximizes US(δ) = Eψ, y[uS(θ, y, δ)]. This is equivalent to maximizing the expected Kullback–Liebler distance between the marginal posterior and prior distributions of θ, and is also equivalent to minimizing the expected entropy of the posterior distribution for θ.
The NSEL utility for θ is given by
(6)
(6) An NSEL-optimal design maximizes the expected utility UV(δ), which is equivalent to minimizing the expectation of the trace of the posterior covariance matrix of θ with respect to the marginal distribution of y.
3.1.1 Evaluating the Expected Utility via Numerical Approximation
For many statistical models, including most nonlinear models, evaluation of uS(θ, y, δ) and uV(θ, y, δ) requires numerical approximation. For given values of y and θ, the components of (Equation5(5)
(5) ) can be approximated as
where
is a size
random sample from the prior distribution of ψ = (θ, γ). These quantities can be incorporated into a nested, or double-loop, Monte Carlo approximation of US(δ):
with {yl, θl}Bl = 1 a sample from the joint distribution of the response and parameters. Intuitively, the “inner sample” of size
is used to approximate the two marginal likelihoods in (Equation5
(5)
(5) ), the first marginal to γ and the second to both γ and θ, and the “outer sample” of size B is then used to approximate the expected utility with respect to the joint distribution of y and θ. This approximation is biased for US(δ) due to the bias in
and
. However, under regularity conditions satisfied by most models of practical importance (Severini Citation2000, pp. 80–81), this bias is of order
(Ryan Citation2003) and hence asymptotically negligible.
For NSEL, E(θw | y, δ) in (Equation6(6)
(6) ) can be approximated via importance sampling:
where
is a random sample from the prior distribution of ψ, and
is the wth element of
. Hence, the following nested Monte Carlo approximation of the expected utility is obtained:
where θlw is the wth element of θl. Here, the inner sample is used to approximate the posterior expectation, and the outer sample used to approximate the expected utility. Importance sampling has commonly been used to estimate posterior quantities for Bayesian design (see Ryan et al. Citation2016 and references therein), although the approximation of the expected utility will again be biased due to bias in
.
In the examples, we set for the evaluation of
in Step 2b of the ACE algorithm (chosen from practical experience). For the comparisons in Steps 2d and 4e, we set
.
3.1.2 Evaluating the Expected Utility via Normal Approximation
The following approximations to US(δ) and UV(δ) are commonly used (Atkinson, Donev, and Tobias Citation2007, chaps. 10, 18), justified via a normal approximation to the posterior distribution of ψ:
with
the Fisher information matrix for ψ, or an approximation thereof, and A = [Ip, 0p × (P − p)]T with Ip the p × p identity matrix and 0a × b the a × b zero matrix. Designs that maximize φS and φV are sometimes referred to as pseudo-Bayesian D- and A-optimal designs, respectively. Note that these expressions also result from taking expectations of the utility functions
which do not depend on y. Unbiased Monte Carlo approximations to φS(δ) and φV(δ) can be obtained via sampling from the prior distribution for ψ:
For comparison of designs, the D-efficiency of design δ1 relative to design δ2 is defined as
(7)
(7)
3.2 Compartmental Model
Compartmental models are applied in pharmacokinetics to study how materials flow through an organism, and have been used extensively to demonstrate optimal design methodology (Atkinson et al. Citation1993; Gotwalt, Jones, and Steinberg Citation2009). The archetypal design problem is to choose n sampling times δ = (t1, …, tn)T, in hours, at which to measure the concentration in a subject of a previously administered drug. Here, concentration is modeled as yi ∼ N(a(θ)μ(θ; ti), σ2b(θ; ti)) , where θ = (θ1, θ2, θ3)T are the parameters of interest, σ2 > 0 is a nuisance parameter, a( · ) and b( · ; ·) are application-dependent functions, and μ(θ; ti) = exp ( − θ1ti) − exp ( − θ2ti).
For this problem, Ryan et al. (Citation2014) assumed that
and found designs using the SIG utility function. Independent log-normal prior distributions were assumed for the elements of θ with, on the log scale, each having common variance 0.05 and expectations log (0.1), log (1), and log (20) for θ1, θ2, and θ3, respectively. These authors also incorporated the constraint max s, t = 1, …, n|ts − tt| ⩾ 0.25, that is, that sampling times must be at least 15 min apart. It is straightforward to incorporate this constraint into design search using the ACE algorithm. In Step 2d,
is maximized over a set
that satisfies the constraint. Phase II of the ACE algorithm is then omitted as replicated sampling times are not permitted.
Ryan et al. (Citation2014) restricted their search for an SIG-optimal design to the class of designs defined via a dimension reduction scheme (DRS) that set the n sampling times to scaled percentiles of a Beta(α1, α2) distribution. Hence, the design problem was reduced to selecting two parameters, α1 and α2. The Müller (Citation1999) simulation algorithm was used to sample from an artificial posterior distribution for α1, α2, with unnormalized density equal to the integrand in (Equation1(1)
(1) ). The chosen design was then the scaled quantiles from the Beta distribution obtained from using the posterior modal values of α1 and α2.
We compare this design with three designs found from ACE: (i) an SIG-optimal design; (ii) a pseudo-Bayesian D-optimal design; and (iii) an optimal choice of α1, α2 for the Beta DRS. For this latter design, the sampling times are given by , with Q(r; α1, α2) the rth quantile of the Beta(α1, α2) distribution. In Step 2d of the ACE algorithm, the sets
and
are given by
For the SIG- and D-optimal designs, and shows trace plots of the approximate expected utility at each iteration of the algorithm. Approximate convergence is demonstrated, for both utility functions, through each run of the algorithm resulting in a similar value of after a relatively small number of iterations. Convergence is, however, achieved more quickly for pseudo-Bayesian D-optimality, which does not require approximation of posterior quantities. This criterion also displays greater consistency in the final approximated expected utility between runs of the algorithm.
Figure 2. (a), (b) Trace plots of for each iteration of the ACE algorithm for SIG and pseudo-Bayesian D-optimality utilities, respectively; in each plot, the black line shows the trace of the expected utility for the best design; (c) designs found from the ACE algorithm: unrestricted SIG-optimal, pseudo-Bayesian D-optimal, Beta DRS SIG-optimal, together with the Ryan et al. (Citation2014) Beta DRS SIG-optimal designs; (d) boxplots for 20 evaluations of
for designs from these four methodologies; (e) approximate expected utility surface for SIG as a function of the Beta DRS parameters; parameter values corresponding to the Ryan et al. (Citation2014) and the ACE DRS designs are marked.
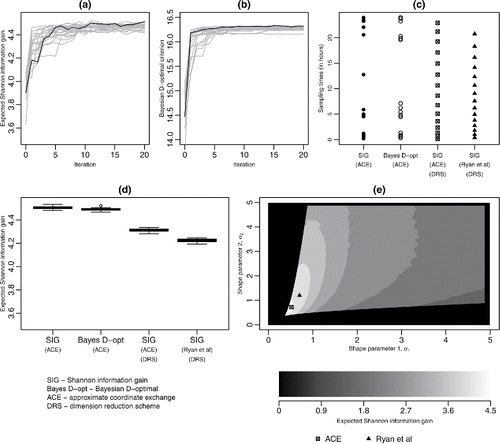
The sampling times for the four designs, shown in , indicate that the designs using dimension-reduction do not display the clustering of points evident in the SIG and pseudo-Bayesian D-optimal designs. The boxplots in , from 20 evaluations of (B = 20,000) for each design, confirm that larger approximate expected utilities are obtained, up to a 5% improvement, when DRS is not used. Here, the pseudo-Bayesian D-optimal design provides a good approximation to the SIG-optimal design.
The DRS design found from ACE outperforms the Ryan et al. (Citation2014) design. To explore this result further, the expected utility surface was investigated as a function of α1 and α2 by sampling 40,000 (α1, α2) pairs from [0, 5]2 and evaluating for each pair. The resulting expected utility surface is shown in , where
for parameter pairs that do not satisfy the 15 min constraint. Both methods identify the relatively small region of high expected utility; the sampling-based algorithm (Müller Citation1999; Ryan et al. Citation2014) fails to identify the optimum point within this region.
3.3 Logistic Regression in Four Factors
Fully-Bayesian design for multi-variable logistic regression has not appeared in the literature, although Hamada et al. (Citation2001) found an SIG-optimal design for a single-variable model and Woods et al. (Citation2006) were the first to find multi-variable pseudo-Bayesian D-optimal designs. Here, we find designs for a first-order logistic regression model in four variables where the response is measured for G groups of ng runs, that is, n = Gng. Let yst ∼ Bernoulli(ρst) be the tth response from the sth group (s = 1, …, G; t = 1, …, ng), with
where
are the parameters of interest, and
(i = s, …, G) are the group-specific nuisance parameters (or “random effects”). Let X = (XT1 ⋅⋅⋅ XGT)T be the n × 5 model matrix where Xs is the ng × 5 matrix with tth row given by xTst. The design matrix D is formed as the last four columns of X, δ = vec(D) has length q = 4n, and
for i = 1, …, q.
The following independent prior distributions for each element of β are assumed:
(8)
(8) We find designs for two different prior distributions for each ωs (s = 1, …, G): (i) a prior point mass at ωs = 0 for all s, resulting in standard logistic regression with homogenous groups; (ii) a hierarchical prior distribution in which the elements of ωs are independent and identically distributed as ωsr ∼ U[ − λr, λr], for r = 0, …, 4, with λr > 0 unknown and having triangular prior density
with (L0, …, L4) = (3, 3, 3, 1, 1).
3.3.1 Logistic Regression With Homogenous Groups
We use ACE to find designs that maximize the SIG and NSEL expected utilities for homogenous logistic regression with ωs = 0 and n = 6, …, 48. For comparison, we also find pseudo-Bayesian D- and A-optimal designs. We also compare to maximin Latin hypercube (LH) designs (Morris and Mitchell Citation1995). For this example, the starting designs for the algorithm were a locally D-optimal design (for SIG and Bayesian D) and a locally A-optimal design (for NSEL and Bayesian A), found from ACE via maximization of ψS(δ) or ψV(δ), respectively, using a point prior distribution for each parameter with support at the mean of each prior distribution in (Equation8(8)
(8) ). presents results (minimum, mean, maximum) of 20 evaluations of (a)
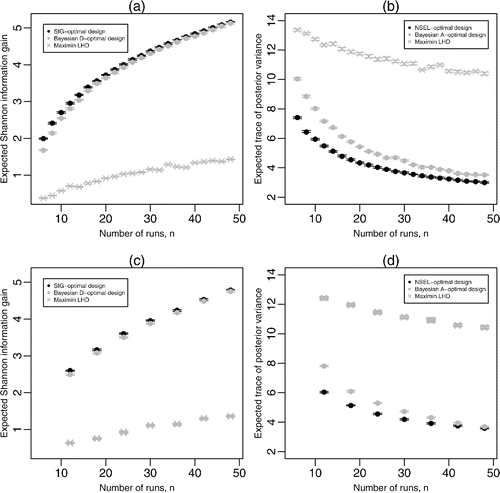
for the SIG-optimal, Bayesian D-optimal, and maximin LH designs, and (b)
for the NSEL-optimal, Bayesian A-optimal, and maximin LH designs, using B = 20,000 Monte Carlo samples. For small n, on average there are substantial differences in expected utility between the fully Bayesian and pseudo-Bayesian designs, with the SIG-optimal design having expected Shannon information gain up to 20% larger than the Bayesian D-optimal design and the NSEL-optimal design having expected trace of the posterior covariance matrix up to 27% smaller than the Bayesian A-optimal design. For both SIG and NSEL, as n increases, the difference in expected utility between these designs and the pseudo-Bayesian designs decreases. For SIG, these findings agree with asymptotic results on the convergence, under certain regularity conditions, of the posterior distribution to a normal distribution (see, e.g., Gelman et al. Citation2014, pp. 585–588). The maximin LH designs, which are model-free space-filling designs, perform poorly under both SIG and NSEL utilities and are not competitive with the model-based designs.
Figure 3. Results from 20 evaluations of (a) for SIG-optimal, pseudo-Bayesian D-optimal, and maximin Latin hypercube designs, and (b)
for NSEL-optimal, pseudo-Bayesian A-optimal, and maxmin Latin hypercube designs, for homogenous logistic regression; (c) and (d) show the same evaluations for hierarchical logistic regression. For the latter two plots, for each value of n, 20 different random assignments are made of the points of the Latin hypercube design to the G groups, and each resulting design is evaluated 20 times. For each design, the central plotting symbol denotes the mean expected Shannon information gain or expected average posterior variance, with the two horizontal lines denoting the minimum and maximum of these quantities.
As there are no comparable results on fully-Bayesian design for multi-variable logistic regression in the literature, we compare the pseudo-Bayesian D-optimal designs for n = 16 and n = 48 found from ACE with designs from the approach of Gotwalt, Jones, and Steinberg (Citation2009). We independently implemented the methodology of these authors to obtain designs for n = 16 and n = 48; we also compare to the n = 16 run design published by Gotwalt, Jones, and Steinberg (Citation2009). For each of these three designs, we calculated the average of D-efficiency (Equation7(7)
(7) ) over 20 Monte Carlo approximations (each with B = 20,000) relative to the appropriately sized design from ACE. The published 16-run design has average efficiency of 82%; the designs from our implementation perform similarly to the ACE designs, with average efficiencies of 99.9% and 101.3% for n = 16 and n = 48, respectively.
3.3.2 Hierarchical Logistic Regression
For hierarchical logistic regression, we again find SIG-optimal and NSEL-optimal designs, along with pseudo-Bayesian D- and A-optimal designs using an approximation to the Fisher information (Pawitan Citation2001, p. 467). We set ng = 6 and G = 2, …, 8, leading to n = 12, 14, …, 48. To reduce the computational burden, B = 1000 was used in Step 4e to find SIG-optimal designs. Previous research has found pseudo-Bayesian D-optimal designs for smaller numbers of variables and group sizes (Waite and Woods Citation2015).
and shows results from 20 evaluations of and
for the SIG-optimal and pseudo-Bayesian D-optimal designs, and the NSEL-optimal and pseudo-Bayesian A-optimal designs, respectively. Again, the performances of maximin LH designs are included for reference (see figure caption for details). A comparison with and , respectively, shows lower expected gains in Shannon information and higher expected posterior variance for the hierarchical logistic regression model due to additional uncertainty introduced by the group-specific parameters. As with designs for homogenous logistic regression, the difference in expected utility between the pseudo-Bayesian designs and the fully-Bayesian designs decreases as n increases, and the LH designs perform poorly.
3.4 Binomial Regression Under Model Uncertainty
Uncertainty over the choice of statistical model π(y, ψ | δ) is common in practice, and has been addressed in pseudo-Bayesian design for generalized linear models by Woods et al. (Citation2006). To demonstrate Bayesian optimal design under model uncertainty, we find follow-up designs for the beetle mortality study of Bliss (Citation1935), a common example used to illustrate binomial regression. In the original dataset, 481 beetles were each administered one of eight different doses (in mg/L) of carbon disulfate. We broadly follow the case study analysis of O’Hagan and Forster (Citation2004, pp. 423–433), who reproduced the data, and assume interest lies in providing a model-averaged posterior distribution of the lethal dose 50 (LD50), the dose required to achieve 50% mortality.
We assume that the binary indicator of death for each beetle is an independent Bernoulli random variable. The number, yk, of deaths from dose xk is modeled as yk ∼ Binomial(nk, ρk), where ρk is the probability of death for the kth dose, which was administered to nk beetles, ∑nk = 1nk = 481. We denote the link function by g(ρk) = ηk, with ηk the linear predictor and consider six models formed by the Cartesian product of three link functions and two linear predictors: the logit, g(ρk) = log {ρk/(1 − ρk)}, the c-log-log, g(ρk) = log { − log (1 − ρk)}, and the probit, g(ρk) = Φ− 1{ρk}, with Φ{ · } the standard normal distribution function; and first-order (ηi = β0 + β1xi) and second-order (ηi = β0 + β1xi + β2x2i) linear predictors.
Let denote the model indicator (see ) and let βu denote the vector of regression parameters under model u. LD50 is then given by
where w = log { − log (0.5)} for the c-log-log link function, and 0 otherwise. We use unit information prior distributions (Ntzoufras, Dellaportas, and Forster Citation2003) for βu | u under each model and set π(u) = 1/6 for u = 1, …, 6. The posterior model probabilities for each model are approximated using importance sampling to evaluate the marginal likelihood of each model, and given in . Samples from the posterior distribution of the model parameters are generated for each of the six models using the Metropolis–Hastings algorithm, and then weighted by π(u | y) to produce a sample from the joint posterior distribution βu, u | y of regression parameters and model indicator. A sample from the model-averaged posterior distribution of LD50 can be obtained by evaluating
for each sampled parameter vector.
Table 1. Approximate posterior model probabilities, π(u | y), for the beetle mortality data.
We consider the design of a follow-up experiment using a further n0 (potentially new) doses. Each dose is to be administered to beetles (k0 = 1, …, n0) and, in each group, the number,
, of beetles that die is recorded. Let y0 be the n0 × 1 vector of the numbers of beetles that die in the follow-up experiment. We assume that
is unknown and adopt a Poisson(λ) prior distribution. Hence
. We choose λ = 60, consistent with the values of nk in the original dataset, and find designs for n0 = 1, …, 10 to estimate the value of LD50 under the NSEL utility function by maximizing
where design δ is the n0 × 1 vector of doses and
is the parameter space for model u. For the purposes of design and modeling, we assume that
for all i = 1, …, n0 but transform the doses to the original scale [1.6907, 1.8839] for the presentation of results.
We can approximate UV(δ) by
where {βul, ul, y0l}Bl = 1 is a random sample from the joint distribution with density π(βu, u, y0 | y), and
where
is a random sample generated from the joint distribution with density π(βu, u | y).
summarizes the results from the ACE algorithm. The doses in the NSEL-optimal design lie in the lower tail of the (original) posterior distribution of LD50 for all values of n0, see . For n0 > 1, the doses are concentrated near a single point (1.77), for example, four replicate points occur for n0 = 10. The approximate expected posterior variance of LD50, , rapidly decreases as n0 is initially increased from 1, see ; the rate of decrease slows as n0 becomes larger.
Figure 4. (a) Posterior density for LD50, the original experimental doses and optimal doses (in mg/L) for each value of n0; (b) boxplots of 20 evaluations of for each n0 for the NSEL-optimal designs; (c) negative approximate expected utility
against dose for n0 = 1; the vertical line indicates δ⋆. (d) negative approximate expected utility
against dose for n0 = 2; ⊠ indicates δ⋆.
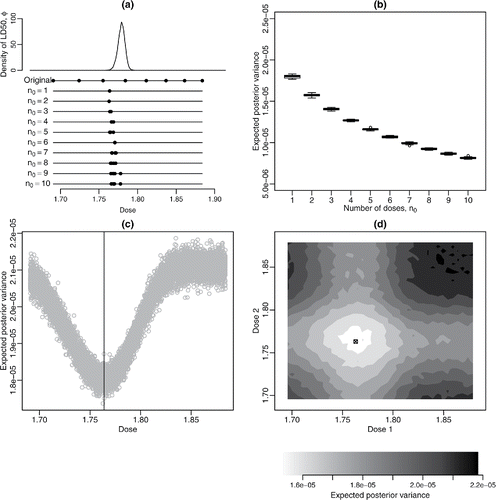
To further investigate the selected designs, the expected utility surface and the performance of the ACE algorithm, we randomly generated 10,000 designs for n0 = 1 and n0 = 2 uniformly from and
, respectively. For each design, we evaluate
and plot against dose; see for n0 = 1 and for n0 = 2. The NSEL design identified by ACE is marked in each plot and, for both values of n0, the minimum negative expected utility is achieved. The variance of the original model-averaged posterior distribution for LD50 is 2.10 × 10− 5. Hence for both n0 = 1 and n0 = 2, it is clear that choosing a design composed of only very high or low doses would have resulted in a negligible expected reduction in variance.
4. Discussion and Future Work
The ACE methodology proposed in this article provides a step-change in the nature and complexity of statistical models and experiments for which Bayesian designs can be obtained. It may be used to find decision-theoretic designs whenever it is possible to simulate values from the prior distribution of the model parameters and responses from the statistical model. The combination of emulating an approximation to the expected utility and the coordinate exchange algorithm has allowed much larger problems to be tackled than was previously possible, both greater numbers of runs and more controllable variables. The algorithm also matches or exceeds the performance of existing approaches for smaller problems, and offers a clear advantage for design selection over the application of a dimension reduction scheme. The new designs made possible by this methodology also allow previously impossible benchmarking of designs from asymptotic approximations.
As presented, ACE can be applied to numerous important practical problems using the available R package. We have applied, or are in the process of applying, ACE to problems from chemical development and biological science. There are also a variety of extensions that could be made to ACE to increase its computational efficiency and applicability. We now highlight a few of these areas.
In ongoing work, we are extending and applying the methodology to find designs for statistical models where the likelihood function is only available numerically as the output from an expensive computer code (see also Huan and Marzouk Citation2013). Such models include those described by the solution to a system of nonlinear differential equations, which are increasingly studied in the field of uncertainty quantification (e.g., Chkrebtii et al. Citation2015).
Convergence of the algorithm may be improved through a reparameterization of the design to remove dependencies between coordinates (e.g., Fletcher Citation1987, p. 19) that can be evident in efficient designs for some models. Such dependencies could be identified through pilot runs of the algorithm or by studying properties of pseudo-Bayesian designs. Additionally, the computational burden of the algorithm could be further reduced by employing alternative approaches to perform each one-dimensional optimization step in the algorithm. For example, a sequential strategy could use an expected improvement criterion modified for stochastic responses (e.g., Pichney et al. Citation2013).
Alternative strategies could also be adopted for the approximation of the expected utility. Zero-variance Monte Carlo (Ripley Citation1987, pp. 129–132, Mira, Solgi, and Imparato Citation2013) could be used to reduce the variance of the Monte Carlo estimator through the introduction of negative correlations via antithetic variables. Combining deterministic approximations, such as expectation propagation, with Monte Carlo methods would remove the need for nested simulation and may work well for nonlinear regression models with normal prior distributions.
Supplementary Material
Download Zip (258.8 KB)Supplementary Materials
The supplementary material includes the designs discussed in the article, and documentation and code to reproduce all the examples. The R package acebayes that implements the ACE algorithm is available on CRAN (https://cran.r-project.org/package=acebayes).
Acknowledgments
This work was supported by the U.K. Engineering and Physical Sciences Research Council through Fellowship EP/J018317/1 for D.C. Woods. The authors thank the editor, an associate editor, and two reviewers for comments that improved the article, and the participants at the “Bayesian Optimal Design of Experiments” workshop (Brisbane, Australia, December 2015; http://www.bode2015.wordpress.com) for useful discussions on extensions and future work. The authors acknowledge the use of the IRIDIS High Performance Computing Facility, and associated support services at the University of Southampton, in the completion of this work.
References
- Amzal, B., Bois, F. Y., Parent, E., and Robert, C. (2006), “Bayesian Optimal Design via Interacting Particle Systems,” Journal of the American Statistical Association, 101, 773–785.
- Atkinson, A., Donev, A., and Tobias, R. (2007), Optimum Experimental Design, with SAS, Oxford: Oxford University Press.
- Atkinson, A. C., Chaloner, K., Herzberg, A. M., and Juritz, J. (1993), “Experimental Designs for Properties of a Compartmental Model,” Biometrics, 49, 325–337.
- Bastos, L. S., and O’Hagan, A. (2009), “Diagnostics for Gaussian Process Emulators,” Technometrics, 51, 425–438.
- Bliss, C. I. (1935), “The Calculation of the Dosage-Mortality Curve,” Annals of Applied Biology, 22, 134–167.
- Chaloner, K., and Larntz, K. (1989), “Optimal Bayesian Design Applied to Logistic Regression Experiments,” Journal of Statistical Planning and Inference, 21, 191–208.
- Chaloner, K., and Verdinelli, I. (1995), “Bayesian Experimental Design: A Review,” Statistical Science, 10, 273–304.
- Chkrebtii, O. A., Campbell, D. A., Calderhead, B., and Girolami, M. (2015), “Bayesian Solution Uncertainty Quantification for Differential Equations” (with discussion), Bayesian Analysis, 11, 1239–1299.
- Dean, A., Morris, M., Stufken, J., and Bingham, D. (eds.) (2015), Handbook of Design and Analysis of Experiments, Boca Raton, FL: CRC Press.
- Fletcher, R. (1987), Practical Methods of Optimization ( 2nd ed.), Chichester: Wiley.
- Gelman, A., Carlin, J., Stern, H., Dunson, D., Vehtari, A., and Rubin, D. (2014), Bayesian Data Analysis ( 3rd ed.), Boca Raton, FL: CRC.
- Gotwalt, C. M., Jones, B. A., and Steinberg, D. M. (2009), “Fast Computation of Designs Robust to Parameter Uncertainty for Nonlinear Settings,” Technometrics, 51, 88–95.
- Hamada, M., Martz, H. F., Reese, C. S., and Wilson, A. G. (2001), “Finding Near-Optimal Bayesian Experimental Designs via Genetic Algorithms,” The American Statistician, 55, 175–181.
- Huan, X., and Marzouk, Y. M. (2013), “Simulation-Based Optimal Bayesian Experimental Design for Nonlinear Systems,” Journal of Computational Physics, 232, 288–317.
- Jones, M., Goldstein, M., Jonathan, P., and Randell, D. (2016), “Bayes Linear Analysis for Bayesian Optimal Experimental Design,” Journal of Statistical Planning and Inference, 171, 115–129.
- Loeppky, J. L., Sacks, J., and Welch, W. J. (2009), “Choosing the Sample Size of a Computer Experiment: A Practical Guide,” Technometrics, 51, 366–376.
- Meyer, R., and Nachtsheim, C. (1995), “The Coordinate Exchange Algorithm for Constructing Exact Optimal Experimental Designs,” Technometrics, 37, 60–69.
- Mira, A., Solgi, R., and Imparato, D. (2013), “Zero Variance Markov Chain Monte Carlo for Bayesian Estimators,” Statistics and Computing, 23, 653–662.
- Morris, M. D., and Mitchell, T. J. (1995), “Exploratory Designs for Computer Experiments,” Journal of Statistical Planning and Inference, 43, 381–402.
- Müller, P. (1999), “Simulation-Based Optimal Design,” in Bayesian Statistics (Vol. 6), eds. J. M. Bernardo, M. J. Bayarri, J. O. Berger, A. P. Dawid, D. Heckerman, and A. F. M. Smith, Oxford, pp. 459–474.
- Müller, P., and Parmigiani, G. (1996), “Optimal Design via Curve Fitting of Monte Carlo Experiments,” Journal of the American Statistical Association, 90, 1322–1330.
- Müller, P., Sanso, B., and De Iorio, M. (2004), “Optimal Bayesian Design by Inhomogeneous Markov Chain Simulation,” Journal of the American Statistical Association, 99, 788–798.
- Ntzoufras, I., Dellaportas, P., and Forster, J. J. (2003), “Bayesian Variable and Link Determination for Generalised Linear Models,” Journal of Statistical Planning and Inference, 111, 165–180.
- O’Hagan, A., and Forster, J. J. (2004), Kendall's Advanced Theory of Statistics, Volume 2B: Bayesian Inference ( 2nd ed.), London: Arnold.
- Overstall, A. M., Woods, D. C., and Adamou, M. (2017), Acebayes: Optimal Bayesian Experimental Design Using the ACE Algorithm, R package version 1.3.
- Pawitan, Y. (2001), In All Likelihood: Statistical Modelling and Inference Using Likelihood, Oxford: Oxford University Press.
- Pichney, V., Ginsbourger, D., Richet, Y., and Caplin, G. (2013), “Quantile-Based Optimization of Noisy Computer Experiments With Tunable Precision” (with discussion), Technometrics, 55, 2–36.
- Rasmussen, C. E., and Williams, C. K. I. (2006), Gaussian Processes for Machine Learning, Cambridge, MA: MIT Press.
- Ripley, B. D. (1987), Stochastic Simulation, New York: Wiley.
- Ryan, E. G., Drovandi, C. C., McGree, J. M., and Pettitt, A. N. (2016), “A Review of Modern Computational Algorithms for Bayesian Optimal Design,” International Statistical Review, 84, 128–154.
- Ryan, E. G., Drovandi, C. C., Thompson, M. H., and Pettitt, A. N. (2014), “Towards Bayesian Experimental Design for Nonlinear Models That Require a Large Number of Sampling Times,” Computational Statistics and Data Analysis, 70, 45–60.
- Ryan, K. J. (2003), “Estimating Expected Information Gains for Experimental Designs With Application to the Random Fatigue-Limit Model,” Journal of Computational and Graphical Statistics, 12, 585– 603.
- Santner, T. J., Williams, B. J., and Notz, W. I. (2003), The Design and Analysis of Computer Experiments, New York: Springer.
- Severini, T. A. (2000), Likelihood Methods in Statistics, Oxford: Oxford University Press.
- Waite, T. W., and Woods, D. C. (2015), “Designs for Generalized Linear Models With Random Block Effects via Information Matrix Approximations,” Biometrika, 102, 677–693.
- Wang, L., and Zhang, L. (2006), “Stochastic Optimization Using Simulated Annealing With Hypothesis Test,” Applied Mathematics and Computation, 174, 1329–1342.
- Weaver, B. P., Williams, B. J., Anderson-Cook, C. M., and Higdon, D. M. (2016), “Computational Enhancements to Bayesian Design of Experiments Using Gaussian Processes,” Bayesian Analysis, 11, 191– 213.
- Woods, D. C., Lewis, S. M., Eccleston, J. A., and Russell, K. G. (2006), “Designs for Generalized Linear Models With Several Variables and Model Uncertainty,” Technometrics, 48, 284– 292.
- Wu, C. F. J., and Hamada, M. (2009), Experiments: Planning, Analysis and Optimization ( 2nd ed.), Hoboken, NJ: Wiley.
