ABSTRACT
Arcobacter has emerged as an important food-borne zoonotic pathogen, causing sometimes serious infections in humans and animals. Newer species of Arcobacter are being incessantly emerging (presently 25 species have been identified) with novel information on the evolutionary mechanisms and genetic diversity among different Arcobacter species. These have been reported from chickens, domestic animals (cattle, pigs, sheep, horses, dogs), reptiles (lizards, snakes and chelonians), meat (poultry, pork, goat, lamb, beef, rabbit), vegetables and from humans in different countries. Arcobacters are implicated as causative agents of diarrhea, mastitis and abortion in animals, while causing bacteremia, endocarditis, peritonitis, gastroenteritis and diarrhea in humans. Three species including A. butzleri, A. cryaerophilus and A. skirrowii are predominantly associated with clinical conditions. Arcobacters are primarily transmitted through contaminated food and water sources. Identification of Arcobacter by biochemical tests is difficult and isolation remains the gold standard method. Current diagnostic advances have provided various molecular methods for efficient detection and differentiation of the Arcobacters at genus and species level. To overcome the emerging antibiotic resistance problem there is an essential need to explore the potential of novel and alternative therapies. Strengthening of the diagnostic aspects is also suggested as in most cases Arcobacters goes unnoticed and hence the exact epidemiological status remains uncertain. This review updates the current knowledge and many aspects of this important food-borne pathogen, namely etiology, evolution and emergence, genetic diversity, epidemiology, the disease in animals and humans, public health concerns, and advances in its diagnosis, prevention and control.
1. Introduction
Food-borne infections are gaining importance due to alarming increase in the number of outbreaks in the recent years throughout the world (Scallan et al. Citation2011; Dhama et al. Citation2013a; Hu et al. Citation2014; Lehmann et al. Citation2015). Important food-borne bacterial pathogens include Escherichia coli, Salmonella, Campylobacter, Yersinia enterocolitica, Staphylococcus, Bacillus cereus, Clostridium perfringens, Cl. botulinum and Arcobacter (Collado & Figueras Citation2011; Calvo et al. Citation2013; Dhama et al. Citation2013a; Hsu & Lee Citation2015; Hänel et al. Citation2016). While the virulence and pathogenesis of certain pathogens has been dissected thoroughly, pathogens like Arcobacter are still in the dark zone needing immediate attention. During recent years, Arcobacter spp. has been identified as an emerging food-borne zoonotic pathogen worldwide and the International Commission on Microbiological Specifications for Foods (ICMSF) has classified Arcobacter as a serious hazard to human health. Arcobacter was very first isolated and described from aborted bovine fetal tissues (Ellis et al. Citation1977) and later from porcine fetuses (Ellis et al. Citation1977, Citation1978; Aydin et al. Citation2007; Rasmussen et al. Citation2013). Arcobacters are implicated as causative agents of diarrhea, mastitis and abortion in animals, while causing bacteremia, endocarditis, peritonitis, gastroenteritis and diarrhea in humans (Jiang et al. Citation2010; Figueras et al. Citation2014; Ferreira et al. Citation2016). Their presence in blood samples of humans with cirrhosis and gangrenous appendicitis (Lau et al. Citation2002) furthermore complicates the situation. Three species of Arcobacter namely A. butzleri, A. cryaerophilus and A. skirrowii are more commonly associated with clinical conditions (Gill Citation1983; Kiehlbauch et al. Citation1991; de Oliveira et al. Citation1997; Wesley Citation1997; Wesley et al. Citation2000; On et al. Citation2002; Collado & Figueras Citation2011; Patyal et al. Citation2011; Bagalakote et al. Citation2014; Ramees et al. Citation2014a, Citation2014b, Citation2014c). As the Arcobacter organisms are commonly isolated from fecal samples of healthy animals and humans and are unable to satisfy the Koch's postulate, their pathology and pathogenicity is under dispute (Shah et al. Citation2011; Ongor et al. Citation2004; Houf & Stephan Citation2007; Figueras et al. Citation2014).
Arcobacters have been isolated from healthy cattle, sheep and pigs (Ho et al. Citation2006a, Citation2006b; De Smet et al. Citation2011; Patyal et al. Citation2011; Ferreira et al. Citation2016). Poultry act as an important reservoir of Arcobacters and as a major source of infection spread (de Boer et al. Citation1996; Lammerding et al. Citation1996; Atabay et al. Citation1998; Manke et al. Citation1998; Johnson & Murano Citation1999a; Wesley & Baetz Citation1999; González et al. Citation2000; Houf et al. Citation2000; Atabay & Aydin Citation2001; Atabay et al. Citation2008; Collado & Figueras Citation2011; Rahimi Citation2014). Recently, the presence of Arcobacter has been reported from a wild bird, namely collared doves (Streptopelia decaocto) (Di Francesco et al. Citation2014). Poultry intestines have been proposed to harbor Arcobacters and contaminate the slaughter houses during processing of the carcasses thereby enhancing the chances of further contamination (Ho et al. Citation2008a). Arcobacters have also been recovered from a variety of foods of animal origin, namely beef, pork, milk, and sea foods, and also from water and vegetables (Son et al. Citation2006; Collado et al. Citation2010; Hausdorf et al. Citation2011; Patyal et al. Citation2011; Shah et al. Citation2012a; Hausdorf et al. Citation2013; Lee & Choi Citation2013; Rahimi et al. Citation2014; Ramees et al. Citation2014a; Lehmann et al. Citation2015; Ferreira et al. Citation2016). Contaminated water and meat play an important role in its transmission (Snelling et al. Citation2006; Collado et al. Citation2010; González & Ferrus Citation2011; Rahimi Citation2014). In abattoir, Arcobacters have been frequently reported from various sources and their persistence has been recorded even after follow up of disinfection procedures, therefore abattoirs act as a potential source of spread, but such full epidemiological information to draw conclusive remark is lacking (Collado et al. Citation2010). Therefore, contamination of food samples and changing trends in human food habits lead to increasing infections of this food-borne pathogen in the human community (Wesley et al. Citation2000; On et al. Citation2002; Patyal et al. Citation2011; Dhama et al. Citation2013a; Rahimi Citation2014; Ramees et al. Citation2014a, Citation2014b, Citation2014c). A higher amount of genetic diversity and increase in antibiotic resistance has been reported among Arcobacter isolates recovered from different parts of the world (Bagalakote et al. Citation2014; Mohan et al. Citation2014; Rahimi Citation2014; Ferreira et al. Citation2016).
Isolation of Arcobacter is considered as gold standard method for diagnostic purposes (Atabay & Corry Citation1998; Aydin et al. Citation2007; Rahimi Citation2014; Ramees et al. Citation2014b). Various types of growth media have been employed for cultural isolation of Arcobacter species (de Boer et al. Citation1996; Atabay & Corry Citation1998; Johnson & Murano Citation1999b; Ohlendorf & Murano Citation2002; Scullion et al. Citation2004; Collado & Figueras Citation2011; Rahimi Citation2014; Ramees et al. Citation2014b); among these most commonly used method is the one which is developed by Houf et al. (Citation2001). Use of membrane filter is considered superior as compared to the use of antibiotics for isolation, since Arcobacters are more sensitive to growth inhibition by antibiotics used in selective media (Atabay & Corry Citation1998; Engberg et al. Citation2000). Despite the availability of numerous isolation techniques, there is no recommended standard method for isolation of Arcobacters (Shah et al. Citation2011). Due to this limiting factor, many of the important cases may go undetected, resulting in underestimation of the prevalence and epidemiological status of Arcobacters (Harrab et al. Citation1998; Calvo et al. Citation2013), and this also complicates the result interpretation as reported in various earlier studies (Brann Citation2001; Prouzet-Mauleon et al. Citation2006).
Arcobacter species are fastidious in nature and are biochemically inert similar to Campylobacter species, which complicates their phenotypic differentiation. Hence, polymerase chain reaction (PCR)-based methods are more commonly used for specific detection and identification purposes (Brightwel et al. Citation2007; Douidah et al. Citation2010), especially the multiplex PCR-based methods targeting genus- and species-specific sequences (Douidah et al. Citation2010; Houf et al. Citation2000; Ramees et al. Citation2014a) along with other advanced diagnostics (Suarez et al. Citation1997; Antolin et al. Citation2001; Moreno et al. Citation2003; González et al. Citation2006; Ramees et al. Citation2014a). Perhaps, conventional PCR is evolving as a more rapid tool for detection of Arcobacter from food and other samples (González et al. Citation2010; de Boer et al. Citation2013). More studies are essential in the field of diagnosis of Arcobacters to improve the sensitivity and specificity of the diagnostic methods that are currently in practice.
In the present review, all the salient aspects on this important food-borne pathogen with main focus on etiological details, evolution, emergence and genetic diversity of different Arcobacter species, prevalence and epidemiology in animals, the disease in general in animals and humans, public health significance, and trends and advances in diagnosis, prevention and control have been emphasized.
2. Etiology
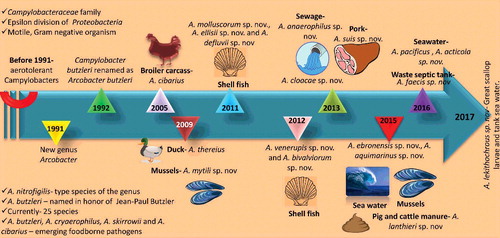
The genus Arcobacter was proposed by Vandamme et al. (Citation1991) and was previously known as ‘aerotolerant campylobacters’ due to their resemblance to the organisms of the genus Campylobacter (Neill et al. Citation1979; Vandamme et al. Citation1991; Aydin et al. Citation2007; Collado et al. Citation2010; Shah et al. Citation2012a; Levican & Figueras Citation2013). Later, it has been revealed that these organisms can be distinguished from Campylobacter by their ability to grow between 15 °C and 30 °C temperature aerobically and need of microaerophilic condition for primary isolation (Collado & Figueras Citation2011; Ferreira et al. Citation2016). Furthermore, based on extensive immunotyping, SDS-PAGE analysis of cellular proteins, fatty acid analysis, DNA–rRNA and DNA hybridization studies, a new genus within the family of Campylobacteriaceae was named as Arcobacter by Vandamme and De Ley (Citation1991). Vandamme et al. (Citation1992b) formally transferred Campylobacter butzleri to the genus Arcobacter as Arcobacter butzleri and also identified five major groups using DNA–DNA hybridization data, these included A. cryaerophilus (two distinct electrophoretic subgroups), A. butzleri, A. nitrophilus and a new species A. skirrowii. The species A. butzleri was named in the honor of Jean-Paul Butzler, a Belgian clinician and microbiologist. The organism has been identified from a wide variety of habitats and hosts which makes the genus atypical within the epsilon subdivision of the Proteobacteria (Debruyne et al. Citation2008).
Arcobacter in Latin means ‘arc-shaped bacterium’, and is included in Campylobacteraceae family consisting of Gram-negative, helical, non-spore forming rods. Together with the genus Helicobacter, Arcobacters form a phylogenetically distinct group referred to either as rRNA superfamily VI or as the epsilon division of the Proteobacteria (Vandamme et al. Citation1991; Lehner et al. Citation2005; Collado & Figueras Citation2011). These bacteria are usually helical, curved or ‘S’-shaped when viewed by conventional light microscopy. Arcobacters are generally 0.2–0.9 μm wide and 1–3 μm long (Wesley Citation1994). These organisms are motile by means of single unsheathed polar flagellum and exhibit a darting corkscrew movement which enables them to pass through membrane filters that retain almost all other bacteria, a feature that is used for their isolation. Biochemically, the majority of Arcobacter species show positive reaction to oxidase, catalase, indoxyl acetate hydrolysis test and nitrate reduction test whereas hippurate hydrolysis test, urease test, and H2S production from triple sugar iron agar (TSI) are negative. The G+C content of their DNA is 27.2–28.2 mol% (Mesbah et al. Citation1989; Vandamme & De Ley Citation1991).
3. Growth parameters
Arcobacter spp. differ physiologically from the Campylobacter spp. as they can tolerate oxygen and can grow at atmospheric temperature below 30 °C (Kjeldgaard et al. Citation2009). A. butzleri strain can grow within a range from 15 °C to 37 °C and with optimal growth at 30 °C (Hilton et al. Citation2001). For primary isolation, microaerobic condition is necessary (Vandamme et al. Citation1991) and no detectable growth is usually seen at 40 °C (Vandamme et al. Citation1992b). It can withstand freezing for up to 6 months at −20 °C and for up to 24 months at −70 °C, but at 55 °C and above the organism gets rapidly inactivated (D'Sa & Harrison Citation2005). Arcobacter strains can tolerate a wide range of pH from 5.5 to 9.5 but optimal growth occurs between pH 6.8–8.0 (Neill et al. Citation1979). Arcobacters have a respiratory type of metabolism and use organic and amino acids as carbon sources. Carbohydrates are neither fermented nor oxidized (Ellis et al. Citation1977). Hitherto studies reported that A. butzlerii was able to grow at 10 °C, the minimum detectable growth temperature for this microorganism (D'Sa & Harrison Citation2005; Kjeldgaard et al. Citation2009) and there was no growth at 40 °C (Hilton et al. Citation2001). Confirmation regarding its tolerance to pH, temperature treatments and salt concentrations has been documented recently (Cervenka Citation2007; Ferreira et al. Citation2016; Salas-Massó et al. Citation2016).
4. Evolution and emergence of Arcobacters
To date, 25 species of Arcobacters have been reported with a significant genetic diversity among and within the species (Collado & Figueras Citation2011; Figueras et al. Citation2012; Levican & Figueras Citation2013; Levican et al. Citation2013a; Sasi Jyothsna et al. Citation2013; Figueras et al. Citation2014; Bagalakote et al. Citation2014; Levican et al. Citation2015; Whiteduck-Le veillée et al. Citation2015; Park et al. Citation2016; Zhang et al. Citation2016). List of Arcobacter spp. and their origin has been presented in . Laishram et al. (Citation2016) for the first time reported the isolation of Arcobacter mytili from India. They also reported that A. butzleri isolated from seafood harbored putative virulence marker genes which are found in human pathogenic strains (Laishram et al. Citation2016). Whiteduck-Léveillée et al. (Citation2016b) identified and characterized the Arcobacter faecis (strain AF1078T), from a human waste septic tank near Ottawa, Ontario, Canada.
Table 1. List of Arcobacter spp. and their origin.
Among these species, A. butzleri, A. cryaerophilus, A. skirrowii and A. cibarius are considered as emerging food-borne pathogens (Ho et al. Citation2006a; Prouzet-Mauleon et al. Citation2006; Snelling et al. Citation2006; Houf & Stephan Citation2007; Collado et al. Citation2009a; Amare et al. Citation2011; McGrego & Wright Citation2015). During the recent years, Arcobacter spp. have gained attention of worldwide researchers as an important emerging food-borne enteropathogen. Among all Arcobacters, A. butzleri is the most common species and has been associated with human disease, such as enteritis, severe diarrhea, bacteremia and septicemia (Engberg et al. Citation2000; Lau et al. Citation2002; Collado & Figueras Citation2011; Collado et al. Citation2014; Figueras et al. Citation2014; Van den Abeele et al. Citation2014; Webb et al. Citation2016). Knowledge gap in the field of emergence and evolution of Arcobacters is very wide as several themes and questions remained unsolved. The genus Arcobacter is evolving rapidly from the day it has been defined. Recently, a symbiotic relationship of Arcobacter with Acanthamoeba castellanii has been identified showing its potential to survive and escape phagocytosis by amoeba (Medina et al. Citation2014). Evolution of this emerging food-borne pathogen needs to be elucidated in a better way so as to find out the epidemiology and virulence factors of this pathogen. Plasmids are usually present in many prokaryotes and they play a major role in evolution of these species (Ricci & Hernandez Citation2000). Recently, several small and a large plasmid were isolated from Arcobacter butzleri. Complete sequence of the large plasmid revealed that putative protein peg21, peg22, peg24, peg25 Peg26 and Peg 29 were showing homology with type IV secretion which plays a role in transfer of DNA material and toxins. This large plasmid can play a role in transfer of genes within Arcobacter spp. leading to evolution of new species (Douidah et al. Citation2014a).
An evolutionary overview of emergence of Arcobacter species with new species additions over the years is presented in .
5. Virulence factors
There is scare research information regarding the virulence genes/factors in Arcobacters and from available studies it is understood that adhesion, invasion of the pathogen, secretion of toxin and pro-inflammatory cytokine (IL-8) plays a major role in establishing infection in the host (Collado & Figueras Citation2011). A. cryaerophilus and A. butzleri are the two main species of Arcobacter studied so far for its adhesive and invasive properties. Studies with rat ileal loop assay showed that former species could cause accumulation of electrolytes and fluids (Ferna´ndez et al. Citation1995). However, the latter one causes intestinal colonization in piglets (Ho et al. Citation2006b). Some studies also reported that Beltsville White turkey was the better animal model for studying the diarrheal infection caused by Arcobacter (Wesley & Baetz Citation1999). Hemagglutinin property has also been reported to be associated with A. butzleri which interacted with the RBCs, hence helps in adhesion of the bacterium (Tsang et al. Citation1996). Impairment of epithelial function was noticed in human HT-29/B6 colonic epithelial monolayers due to A. butzleri infection. Claudin protein family is most important to the barrier function and claudin-1, 5 and 8 have barrier sealing properties (Bucker et al. Citation2009). Tight junction proteins namely claudin-1, -5 and -8 have not been found to be expressed properly during A. butzleri infection in human colonic epithelial cells, which leads to epithelial barrier dysfunction (Bucker et al. Citation2009). Similar effect of claudin-1 dysfunction was noticed in enteropathogenic E. coli-infected human intestinal T84 monolayers (Bucker et al. Citation2009). Like Campylobacter and Helicobacter, the release of IL-8 in Arcobacter infection may play a role in causing diarrhea (Ho et al. Citation2007; Ferreira et al. Citation2014b). Release of pro-inflammatory cytokines is a major virulence factor for H. pylori and Campylobacter, similarly Arcobacter leads to release of pro-inflammatory cytokines (Collado & Figueras Citation2011). Zur Bruegge et al. (Citation2014) reported that Arcobacter has less potential for intracellular survival and also documented strain specific differences in invasion and survival capacities of the organism inside the host. Arcobacter isolates when cultured in Vero cells produced cell elongation, which showed their ability to produce enterotoxin and some isolates along with this ability produced vacuoles in cells showing that these produce vacuolizing toxin (Villarruel-López et al. Citation2003). Putative role played by lipopolysaccharide (LPS) and the composition of LPS have not been elucidated properly in Arcobacter infection. Lipooligosaccharide (LOS) has been characterized in a marine Arcobacter (A. halophilus) and LPS has been reported to play a role in the survival of this organism in marine environment (Silipo et al. Citation2010). As Arcobacter is motile like Campylobacter, their flagella may also have a role in invasion of cells as reported in other bacteria, wherein flagella helps in motility and also in colonization of the pathogen (Ho et al. Citation2008b). Studies regarding the role played by the host immune mechanisms need to be focused for exploring virulence mechanisms of Arcobacters as host can also participate during infection and some of the host immune mechanisms can even further aggravate diarrhea (Collado & Figueras Citation2011).
Ten putative genes namely cadF, ciaB, cj1349, hecA, iroE, hecB, irgA, mviN, pldA and tlyA have been recognized in A. butzleri ATCC 49616 genome (Miller et al. Citation2007; Douidah et al. Citation2012; Ferreira et al. Citation2014b). Functions of the different genes include: cadF gene and cj1349 gene code for outer membrane proteins facilitating intestinal epithelial cell to cell contact by adhering to fibronectin; ciaB gene is involved in invasion of host cell, pldA gene codes for outer membrane phospholipase A which hydrolyzes acyl ester bonds, tlyA gene is a hemolysin gene, irgA gene codes for an outer membrane receptor for enterobactin, hecA gene is a member of filamentous hemagglutinin family and hecB gene codes for hemolysin activation protein (Douidah et al. Citation2012). A study conducted by Tabatabaei et al. (Citation2014) in Southern Iran revealed six virulence genes (cadF, ciaB, cj1349, mviN, pldA and tlyA) in Arcobacter spp. These six genes were present in all the A. butzleri isolates. The cadF, ciaB, cj1349, mviNin, pldA and tlyA genes were present in 55% and 53%, 98% and 87%, 45% and 60%, 90% and 80%, 33% and 13%, and 38% and 40% of A. cryaerophilus and A. skirrowii, respectively. Identification of virulence genes revealed the pathogenic nature of the isolated species of Arcobacter. It was well proved that various virulence genes were responsible for adhesion namely CadF, HecA and Cj1349, invasion CiaB, lysis of erythrocytes HecB, TlyA and PldA, iron acquisition and maintaining of infection IrgA and IroE, and peptidoglycan biosynthesis MviN in other bacteria (Ruiz Citation2008; Flanagan et al. Citation2009). However, it is unknown whether these putative virulence genes show similar actions in Arcobacter. Even though A. butzleri has been shown to adhere and invade into various cell lines in vitro, no correlation have been shown between A. butzleri virulence genes and adhesive and invasive phenotypes (Karadas et al. Citation2013; Levican et al. Citation2013b). Similarly, presence of six virulence genes in the Arcobacter spp. isolates was also reported by Karadas et al. (Citation2013) and Lehmann et al. (Citation2015). Likewise, five virulence genes (ciaB, cadF, cj1349, irgA and hecA) were detected from Arcobacter isolated from different sources namely meat, shellfish, sewage, feces from pigs, sheep and chickens, environmental sources, piggery effluent, seawater roots of Spartina alterni flora and porcine aborted fetus from Nottingham, United Kingdom (Levican et al. Citation2013b). Draft genome of Arcobacter from various sources like pig, dairy cattle manure origin, broiler carcasses and human feces have been recently documented which can help in exploring the various genes and their roles in virulence and pathogenicity (Adam et al. Citation2014a, Citation2014b, Citation2014c). Whiteduck-Léveillée et al. (Citation2016a) reported the prevalence of virulence-associated genes (VAGs) namely ciaB (90%), mviN (70%), tlyA (50%) and pldA (45%) genes were significantly higher when compared to hecA (16%), hecB (10%) and each of irgA and cj1349 (6%) genes, respectively in A. butzleri, A. cryaerophilus and A. skirrowii.
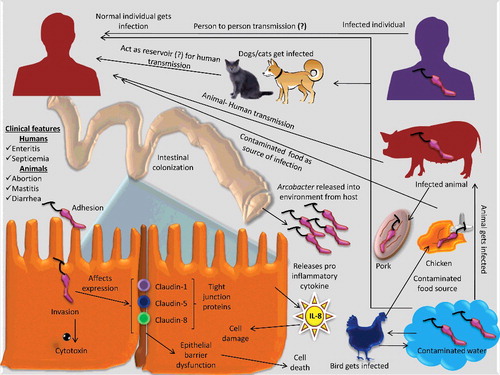
An overview of pathogenesis, mechanisms of virulence, transmission routes and ways/means of release of Arcobacters entering the human food chain is presented in .
Figure 2. Pathogenesis and transmission of Arcobacter. Arcobacter after entry through the digestive tract, colonize the intestine. Several mechanisms like production of cytotoxin, affecting expression of tight junction proteins, release of pro-inflammatory cytokines and epithelial barrier dysfunction lead to cell death. The organism is released into the environment where it contaminates water and food sources thereby entering the human food chain.
6. Epidemiology and transmission in animals
Arcobacters have been reported worldwide from chickens, domestic animals (cattle, pigs, sheep, horses, dogs), reptiles (lizards, snakes and chelonians), meat (poultry, pork, goat, lamb, beef, rabbit), vegetables and from humans in different countries like Belgium, United States of America, Denmark, Brazil, Australia, Italy, the Netherlands, Malaysia, Japan, Spain, Czech Republic, Korea, Egypt and India (Lehner et al. Citation2005; Atabay et al. Citation2006; Ho et al. Citation2006a; Snelling et al. Citation2006; Pejchalova et al. Citation2008; Collado et al. Citation2010; Lee et al. Citation2010; Vilar et al. Citation2010; Amare et al. Citation2011; González & Ferrús Citation2011; Patyal Citation2011; Suelam Citation2012; Ramees et al. Citation2014a, Citation2014b, Citation2014c; Mohan et al. Citation2014; Gilbert et al. Citation2014). Arcobacter is a potential food- and water-borne pathogen, and thus pose serious public health concerns (González et al. Citation2007b; Gugliandolo et al. Citation2008; Miller et al. Citation2009; Ferreira et al. Citation2016). Among the 25 recognized species till date, A. butzleri, A. cryaerophilus and A. skirrowii are the species of veterinary importance. Different Arcobacter species have been isolated from a range of animals and their discharges such as saliva, feces, and from the vagina (Kabeya et al. Citation2003a; Ho et al. Citation2006a). Isolation of Arcobacter from feces of healthy pigs and also from dogs has been documented. In dogs, A. cryaerophilus was more commonly recovered (Ho et al. Citation2006b; Houf et al. Citation2008). Various recovery rates have been documented from fecal samples of different species. From India, Patyal et al. (Citation2011) recorded a prevalence rate of 21% in pigs, 15% in poultry droppings and 3% in human diarrheal samples; Mohan et al. (Citation2014) recorded isolation rate of 8% in poultry droppings, 10% from bovine fecal samples, 12% from pigs and 2% in human diarrheal samples; while, Ramees et al. (Citation2014a) recorded an isolation rate of 2% from human diarrheal samples. From Belgium, Van Driessche et al. (Citation2003) recorded Arcobacter isolation of 44% (pigs), 39% (bovine), 16% (ovine) and 15% (equine) in fecal samples collected at slaughterhouses. A. thereius has also been isolated from liver and kidneys of pigs aborted spontaneously but its exact pathogenic role could not be elucidated (Houf et al. Citation2009). Feco-oral route of transmission play an important role in establishing infection to cattle (Shah et al. Citation2013; Scarano et al. Citation2014). Shirzad Aski et al. (Citation2016) reported the occurrence of Arcobacter spp. in cattle and sheep from Southern Iran as 9% (27/308) by cultural isolation and 14% (44/308) by PCR. The predominant species was A. butzleri in both cattle (58%) and sheep (55%). These results suggested that cattle and sheep are significant intestinal carriers for Arcobacter spp. Very recently, water contamination of Arcobacter (80%) in a farm was documented with A. skirrowii (13%), A. butzleri (40%) and A. cryaerophilus (63%). Same Arcobacter species were isolated after two months from the water troughs which indicates the capacity to form biofilms or as a secondary bioformer (Giacometti et al. Citation2015). This study also reported the presence of A. butzleri in oral cavity of cats. Similar reports regarding the presence of two different Arcobacter spp. (A. butzleri and A. cryaerophilus) from the oral cavity of cats have been reported earlier with 79% positive cases of Arcobacter by using PCR as a diagnostic tool (Fera et al. Citation2009). Hence, pet animals may act as a carrier and can play a part in transmission of Arcobacter to humans.
Prevalence rate of Arcobacter spp. from healthy cattle has been reported to vary in different parts of the world indicating its presence without any clinical manifestation and suggest that cattle can act as a reservoir. shows the prevalence of Arcobacter spp. in healthy cattle from different countries of the world. A. butzleri was the most common organism isolated in dairy farms when compared to other Arcobacter species which may be attributed to the ability of this organism to thrive in different environmental conditions (Golla et al. Citation2002; Giacometti et al. Citation2015). Merga et al. (Citation2011, Citation2013) reported prevalence of 43% from feces of cattle and 40% from feces of sheep in United Kingdom. A study in Belgium reported prevalence of 59% to 85% Arcobacter from fecal samples of pigs (Van Driessche et al. Citation2004). Arcobacter can colonize the gall bladder which shows its potential to cause liver and bile duct infections (Aydin et al. Citation2007). Both vertical and horizontal transmissions of Arcobacters have been documented in animals. Their transplacental transmission has also been documented in pigs (Ho et al. Citation2006b).
Table 2. Prevalence of Arcobacter spp. in healthy cattle from different countries of the world.
Arcobacter has also been reported from monkey's feces (14%) in Canada (Higgins et al. Citation1999; Shah et al. Citation2011). A. butzleri has been the only Arcobacter spp. isolated from both diarrheic as well as healthy non-human primates (Wesley et al. Citation2003; Stirling et al. Citation2008). Microscopic examination of the intestines from these non-human primates revealed chronic active colitis (Anderson et al. Citation1993). Reports of Arcobacter spp. from raccoons (Procyon lotor) was documented for the first time in the year 2004, an issue of public health importance as raccoon share living environment with humans in several parts of the world (Hamir et al. Citation2004). Other species like Galapagos turtle, white Rhinoceros, Gazelle, Rhea and Alpaca are all in the list of animals infected by Arcobacter spp. (Shah et al. Citation2011).
Collado et al. (Citation2014) reported the prevalence of Arcobacter from different marine species like surf clams (88%), where isolation was highest, followed by razor clams (65%), mussels (33%), clams (24%), scallops (18%) and oysters (15%). A moderate rate of prevalence has been reported from sea food (21%) from India (Patyal et al. Citation2011). A. butzleri strains had the ability to survive inside the amoebae known as Acanthamoeba castellanii for the period of 240 h, which suggest that A. butzleri was resistant to amoebic digestion. Moreover, A. castellanii could act as an environmental reservoir and transmission vehicle for Arcobacter (Villanueva et al. Citation2016).
Poultry act as potential reservoirs for Arcobacters and these organisms have been recovered from live poultry skin, droppings and meat (Corry & Atabay Citation2001; Kabeya et al. Citation2004; Lehner et al. Citation2005; Ho et al. Citation2008b; Collado et al. Citation2009a; Amare et al. Citation2011; Collado & Figueras Citation2011; Ramees et al. Citation2014a; Rahimi et al. 2014). A study by Giacometti et al. (Citation2015) indicated absence of Arcobacter spp. in pigeon gut samples (47 numbers tested) and that these birds do not have any role in transmission of Arcobacter to cattle. Various studies have documented the isolation of Arcobacters from feces of broilers with different percentages like 12% in Costa Rica, 14% in Japan and 1.3% from Nigeria (Kabeya et al. Citation2003a; Bogantes et al. Citation2015). Bogantes et al. (Citation2015) also reported 20%, 36% and 12% isolation of Arcobacter from feces of geese, duck and layers. Atabay et al. (Citation2008) reported 18% prevalence of Arcobacter from geese in Turkey.
7. Pathogenesis of Arcobacter spp.
Presently, the pathogenic potential of Arcobacter remains unclear. Only few studies have attempted to explore the pathogenic mechanisms associated with Arcobacter. In vitro cell culture studies of animal and human origin revealed that Arcobacter spp. can adhere to eukaryotic cells, invade and produce toxins resulting in damage to host cells. Adhesion and invasion of bacteria to host cells are essential for successful colonization and establishment of infection (Kolling et al. Citation2012), which might be responsible for potential pathogenic nature of various Arcobacter spp. The mechanism responsible for A. butzleri induced diarrhea was explored by Bucker et al. (Citation2009), wherein it was shown to be related to functional impairment of the epithelial barrier in human colonic carcinoma cells (HT-29/B6), by enhancing the macromolecular permeability through paracellular pathway and decreasing the epithelial resistance. Alteration in the tight junction protein composition and distribution was reported during Arcobacter infection, with decreased expression of sealing-associated proteins, namely claudin-1, claudin-5 and claudin-8, which might be responsible for diarrhea induced by A. butzleri infection through leak flux mechanism.
8. Disease in animals caused by Arcobacters
Most of the infections caused by Arcobacter remain asymptomatic in nature and only few set up clinical condition. Arcobacter in animals cause abortions, diarrhea and mastitis (Logan et al. Citation1982; Neill et al. Citation1985; Vandamme et al. Citation1992a; On et al. Citation2002; Aydin et al. Citation2007; Collado & Figueras Citation2011). In pigs, cattle, and horses, A. butzleri cause enteritis and/or diarrhea, while in sheep and cattle A. skirrowii cause diarrhea and/or hemorrhagic colitis (Ho et al. Citation2006b, Vandamme et al. Citation1992b). A. cryophillus is the predominant species isolated from aborted animals, but it has also been isolated from preputial sheath washings of healthy bovine (Gill Citation1983) and from vaginal swabs of cows with no clinical disease (Kabeya et al. Citation2003a). It has also been isolated from feces of healthy animals such as pigs and cattle, thus raising doubt on its pathogenicity (Kabeya et al. Citation2003a; Van Driessche et al. Citation2003, Citation2004, Citation2005; Ongor et al. Citation2004; Aydin et al. Citation2007). To substantiate, A. cibarius, A. butzleri and A. cryaerophilus were recovered from piggery effluent and also from soil being irrigated by the effluent (Chinivasagam et al. Citation2007). A. cryaerophilus has been associated with porcine abortion, sows with reproductive problems, and preputial fluid of boars and fattening pigs (Neill et al. Citation1985; de Oliveira et al. Citation1997). A. butzleri and A. skirrowii have been less frequently reported from aborted animals (de Oliveira et al. Citation1997; Collado & Figueras Citation2011). Recently, reports have been documented in South Africa regarding the involvement of A. skirrowii induced abortions in ewes. This report further highlighted the need to get insight into the ability of this pathogen to cause abortion in other animals (Bath et al. Citation2013). However, pathology and pathogenesis of Arcobacter infection in animals need to be elucidated properly by unrevealing the pathogenesis of these organisms in different animal models and clinical experiments involving both in vitro and in vivo studies, the better understanding of which could pave the way for developing suitable control strategies and counter this important emerging pathogen.
Although reports from animals are restricted to terrestrial animals, one study reported the isolation of A. cryaerophilus from a natural infection of a fish namely rainbow trout (Oncorhynchus mykiss). The in vivo pathogenicity experiment of the isolated strain revealed infection of intestine, liver and kidney and also caused death of the infected fish (Yildiz & Aydin Citation2006; Collado & Figueras Citation2011). So far, laboratory animal models for studying A. butzleri pathogenesis are lacking because its pathogenesis depends on species and breed of the host. Similarly, A. butzleri did not induce disease in conventional chicken, but can cause disease in turkey strains (Wesley & Baetz Citation1999). Similarly, experimental infection of adult rats with Arcobacter led to watery diarrhea and imbalanced serum electrolytes which depends on the bacterial load, while neonatal albino rats suffered from self-limiting diarrhea, small intestinal and hepatic necrosis (Adesiji Citation2010; Adesiji et al. Citation2012). Açik et al. (Citation2016) for the first time experimentally studied A. butzleri infection in wild zebrafish. The zebrafish were inoculated with mean infective dose (ID50) of A. butzleri as 1.3 × 108 CFU/mL by immersion and 1 × 105 CFU/mL by intraperitoneal injection. Histopathological examination revealed acute inflammation characterized by neutrophils and plasma cells, local necrosis and congestion in various visceral organs. These findings suggested that zebrafish could be used as a laboratory animal model to study the pathogenicity of Arcobacters.
Immunological parameters after Arcobacter infection in a murine model have been investigated. After oral administration of A. butzleri, the bacteria readily colonize the murine intestinal tract. However, infected mice did not show any overt clinical signs, the bacteria not only induced intestinal immune responses but also systemic immune responses in extra-intestinal places depending on the strain and time course of infection (Gölz et al. Citation2015a; Heimesaat et al. Citation2015a). These immune responses were TLR-4 dependent recognized by LOS and LPS derived from the cell walls of Gram-negative bacteria in gnotobiotic IL-10–/– mice lacking TLR-4 with less pronounced intestinal and systemic immune responses (Gölz et al. Citation2015b; Heimesaat et al. Citation2015b; Heimesaat et al. Citation2016). The mucin-2 is a pivotal constituent of the intestinal mucosa providing first line defense against intestinal pathogens (Strugala et al. Citation2003) and was downregulated in the colon, but not in the ileum of A. butzleri infected mice (Gölz et al. Citation2016b; Heimesaat et al. Citation2016). A. butzleri induces antimicrobial host immunity and cytokines expression such as IL-17A, IL-22, IFN-γ and their master regulator IL-23 in intestine that are secreted by T helper (Th) 17 cells. During A. butzleri infection in murine, IL-22 and IL-18 in intestine were upregulated in a strain-dependent manner indicating that the IL-22/IL-18 axis might be responsible for regulating the immune responses during arcobacteriosis (Gölz et al. Citation2016a,Citation2016b; Heimesaat et al. Citation2016). A. butzleri infection in gnotobiotic IL-10–/– mice showed up-regulation of mRNA levels of pro-inflammatory cytokines such as TNF and IL-1β, and MMP-2 and MMP-9 in the intestine (Heimesaat et al. Citation2016).
9. Human infections with Arcobacters
A. cryaerophilus was the first Arcobacter species to be identified from humans in the year 1988 (Tee et al. Citation1988; Collado et al. Citation2011). Arcobacter has been isolated from feces and blood samples of humans and symptoms may range from diarrhea to septicemia (Fisher et al. Citation2014). Gastrointestinal manifestations are the common signs in humans and may be exhibited as watery diarrhea in case of A. butzleri infection, while bloody diarrhea is usually noticed in Campylobacter jejuni (Collado et al. Citation2011). Several studies indicate the prevalence of Arcobacter in humans from different countries of the world. Arcobacter spp. has also been attributed to one of the bacterial agents of traveler's diarrhea along with E. coli, Shigella, Salmonella and Campylobacter (McGrego & Wright Citation2015). Recently, an invasive form of Arcobacter infection in an 85-year-old immunocompromised man was documented (Arguello et al. Citation2015).
9.1. Ways of transmission in humans
Arcobacter in humans mainly causes enteric problem and there are several modes of transmission by which the pathogen can colonize the host cells (Collado & Figueras Citation2011). Transmission route can be feco-oral or direct from animals (Jacob et al. Citation1998; Shah et al. Citation2011; Ferreira et al. Citation2016). Arcobacters are mainly transmitted through contaminated food (vegetables, chicken and pork meat) and water sources which may be contaminated through sewage. Several reports regarding the presence of Arcobacter in water have been documented and hence consumption of contaminated water acts as an efficient source of infection (Jacob et al. Citation1993; González et al. Citation2010; Collado et al. Citation2010; Lee & Choi Citation2013). Sewage contamination is another possible way of infection (Collado et al. Citation2010; Merga et al. Citation2014). Among the bacterial population in sewage, Arcobacter shares in the region of 5%–11% (Fisher et al. Citation2014). Chicken meat, pork and sea foods which are consumed uncooked or partially cooked can lead to infection as these are the major source where Arcobacter was isolated by several workers (Collado et al. Citation2009a).
9.2. Food-borne transmission and prevalences
Arcobacters have been isolated from a variety of meat samples of chicken, pork, beef and seafood (Colado & Figueras Citation2011). Chicken meat has been reported with highest prevalence for Arcobacter followed by pork and beef (Shah et al. Citation2011). Different prevalence rates have been recorded in chicken meat samples in India ranging from 12% to 33% (Patyal et al. Citation2011; Mohan et al. Citation2014; Ramees et al. Citation2014b). Recently, different Arcobacter spp. were isolated (22 samples (11%) of the 200 samples examined) from poultry in Costa Rica (Bogantes et al. Citation2015). Highest incidence of A. butzleri was reported with 83% from chicken meat followed by beef (20%) and pork (15%) in Poland (Zacharow et al. Citation2015). Villalobos et al. (Citation2013) reported isolation of Arcobacter as 17% from poultry visceral samples sold in Costa Rica and most of these isolates were resistant to chloramphenicol and ampicillin. Badilla-Ramírez et al. (Citation2016) reported that A. butzleri had the capacity to grow at 4 °C and 10 °C. They suggested that A. butzleri is a powerful bacterium for surviving in various storage temperatures, and is a potential health risk for poultry meat consumers. A study conducted in Germany by Lehmann et al. (Citation2015) reported the prevalence of Arcobacter spp. as 34% from fish meat, 27% from poultry meat and 2% from minced meat. Similarly, De Smet et al. (Citation2010) reported the presence of Arcobacter from pre- and post-chilled bovine carcasses indicating the need for hygienic practices to break the transmission cycle. At various time period reports of Arcobacter identification from different meat sources around the world has been documented (Vytrasová et al. Citation2003; Rivas et al. Citation2004; Van Driessche & Houf Citation2007a). Several contraindications remain in the isolation of Arcobacter from intestine of poultry carcasses during slaughter house processing though it is believed that time and way of sample collection have a positive role in result interpretation (Houf et al. Citation2002a; Van Driessche & Houf Citation2007b). Water treatment plant and the environment of the slaughter house were implicated for the contamination of poultry carcasses with Arcobacter in Iran (Khoshbakht et al. Citation2014). Arcobacter was isolated from rabbit meat in Egypt (Suelam Citation2012) and also from Spain (Collado et al. Citation2009a). Four Arcobacter spp. (A. butzleri, A. cryaerophilus, A. skirrowii and A. cibarius) have been isolated in high numbers from meat, especially from chicken carcasses. Isolation of Arcobacter from sea foods like oysters (Romero et al. Citation2002), fishes (Patyal et al. Citation2011), shellfish and clams (Collado et al. Citation2009a) are also available.
Reports of Arcobacter isolation from milk samples are also available from different parts of the world, indicating the possible role of milk in its transmission. In a study from Selangor, Malaysia, 5.8% prevalence of Arcobacter spp. from cow milk samples, wherein A. butzleri was the foremost species (60%), followed by A. cryaerophilus (40%), but goat samples were found negative for Arcobacter was reported (Shah et al. Citation2012a). Similarly, Ertas et al. (Citation2010) reported 6% prevalence from raw milk samples in Turkey and Revez et al. (Citation2013) reported 15% prevalence in Finland. Study conducted by Giacometti et al. (Citation2014) showed that A. butzleri and A. cryaerophilus survived for a period of six days in milk stored at 4 °C and 10 °C. Hence, it was concluded that milk can serve as an efficient source of Arcobacter infection to humans. The organism also survived during processing and storage of water buffalo mozzarella cheese, fresh village cheese and sheep ricotta cheese (Serraino et al. Citation2013a; Yesilmen et al. Citation2014; Scarano et al. Citation2014). Studies showed that Arcobacter was also isolated from inline milk filters used in farms (Serraino et al. Citation2013b). Fecal shedding of the organism by cattle can lead to contamination of milk and beef carcasses leading to infection of humans (Shah et al. Citation2012b). Several outbreaks of human gastroenteritis associated with Arcobacter have been linked to the consumption of contaminated fresh vegetables and thus could act as an important source of infection (González & Ferrus Citation2011: Lee & Choi Citation2013). González et al. (Citation2010) reported the presence of A. butzleri in 10 out of 50 fresh lettuce samples screened (20%). Recently, some Arcobacter species were reported on lettuce (González & Ferrús Citation2011) and from carrot-processing plant (Hausdorf et al. Citation2011). Mottola et al. (Citation2016) reported that the occurrence of Arcobacter spp. on pre-cut ready-to-eat (RTE) vegetables was 28% (44/160), of which 91% (40/44) isolates were A. butzleri and 9.1% (4/44) isolates were A. cryaerophilus. The study also revealed widespread distribution of VAGs among the Arcobacter isolates tested. These results indicated that the possibility of health risks associated with the direct consumption of raw vegetables. Food-borne infection due to Arcobacter spp. was reported from Wisconsin, USA, where attendees of a wedding reception who consumed roasted chicken suffered from the illness. PCR and sequence analysis confirmed two species of Arcobacter namely A. butzleri and A. cryaerophilus from the patients (Lappi et al. Citation2013). Person-to-person transmission of A. butzleri was reported in an Italian school with symptoms of recurrent abdominal cramps (Vandamme et al. Citation1992a; González & Ferrús Citation2011).
9.3. Water-borne transmission and prevalences
Contaminated water is considered as an important source of Arcobacter infection to human beings (Jacob et al. Citation1998; Ho et al. Citation2006a; Collado et al. Citation2010). It has been estimated that 63% of A. butzleri infections are acquired in humans by the consumption of or contact with contaminated water (De Smet et al. Citation2011). Arcobacters have been recovered from rivers, lakes, groundwater and seawater, as well as from plankton (Rice et al. Citation1999; Moreno et al. Citation2004; Collado et al. Citation2008; Fera et al. Citation2010). A 4% prevalence of Arcobacter was recorded among 175 samples of drinking water collected at Kayseri, Turkey (Ertas et al. Citation2010). Several water-borne Arcobacter outbreaks associated with drinking water have been documented including those reported from Idaho, USA (Prouet-Mauleon et al. Citation2006), Slovenia (Kopilovic et al. Citation2008) and Ohio, USA (Fong et al. Citation2007). Same species of Arcobacters were detected from drinking water treatment plants and infected humans in Germany showing that water acts as an important source of infection (Jacob et al. Citation1993; Jacob et al. Citation1998; Collado & Figueras Citation2011). Seasonal variations in the prevalence rate of Arcobacter in water treatment plant in Catalonia, Spain, has also been reported with 92% prevalence in spring, 83% in summer and 75% in winter (Collado et al. Citation2010). Reports regarding isolation of Arcobacters from sea water have also been presented (Ansari et al. Citation2015; Levican et al. Citation2015). In this regard, contradictory findings were also stated by Ottaviani et al. (Citation2013) where they found that A. butzleri can neither grow experimentally in sea water nor bioaccumualte in mussels. Levican et al. (Citation2014) found that prevalence of the organism in water sources increased in summer period as the water temperature increased. An interesting study with dishcloth showed the presence of various ciliates along with other food borne pathogen including Arcobacter spp. which urge for adopting good hygienic practices while cooking (Chavatte et al. Citation2014). Talay et al. (Citation2016) reported that 36% (41/115) of water samples from various sources (66 sewages, 25 rivers, 16 spring waters and 8 drinking waters) were showed positive for Arcobacter spp. in Izmir, Turkey. Among them, A. butzleri was reported in 34% of samples (39/115; 24 sewages, 13 rivers and 2 spring waters). The occurrence of Arcobacter spp. in finfish was 19% (8/42), shellfish 15% (5/34) and 21% (5/24) from water samples (Laishram et al. Citation2016). These results indicated that environmental water samples are common sources for Arcobacter spp.
9.4. Disease in humans and public health concerns
A study in South Africa showed A. butzleri to be the third most prevalent species in feces of man while in Belgium and France it was reported to be the fourth most prevalent species (Van Driessche et al. Citation2003; Prouzet-Mauleon et al. Citation2006; Samie et al. Citation2007; Van den Abeele et al. Citation2014). Acrobacter spp. has been associated with enteritis and bacteremia in humans, but still it has not been recognized as a potential human pathogen (Hsueh et al. Citation1997; Engberg et al. Citation2000; Wybo et al. Citation2004; Figueras et al. Citation2014). Arcobacter are also detected in blood samples of patients with clinical conditions like liver cirrhosis and appendiculitis (Yan et al. Citation2000; Lau et al. Citation2002; Aydin et al. Citation2007). The enteritis caused by Arcobacter is an acute diarrhea lasting for 3–15 days, sometimes becoming persistent or recurrent for more than two weeks or even as long as two months (Vandenberg et al. Citation2004). The condition is often accompanied by abdominal pain and nausea and some patients also experience fever, chills, vomiting and weakness (Vandamme et al. Citation1992a).
A. butzleri was frequently isolated more from diarrheic cases as compared to non-diarrheic patients. With a prevalence of 8%, A. butzleri was found to be the etiological agent of traveller's diarrhea acquired by USA and European travellers to Guatemala, Mexico and India (Jaing et al. Citation2010; Shah et al. Citation2011; McGregor & Wright Citation2015). A. butzleri shows similar features of C. jejuni with watery diarrhea. But the diarrhea associated with A. butzleri seems more persistent and is less acute than that caused by C. jejuni (Vandenberg et al. Citation2004). Lerner et al. (Citation1994) isolated A. butzleri from the stool samples of two patients with persistent diarrhea and severe abdominal cramps. A. butzleri was the only potential enteric pathogen isolated during an outbreak among nursery and primary school children in Italy, in this case, 10 infected children suffered from recurrent abdominal cramps without diarrhea (Vandamme et al. Citation1992a). A study conducted on the prevalence of Helicobacter, Campylobacter, and Arcobacter with 322 stool specimens from patients with and without HIV revealed that A. butzleri was the third most prevalent species after H. pylori and C. jejuni in South Africa (Samie et al. Citation2007). A. butzleri is considered as the fourth most common Campylobacter-like organism next to Campylobacter jejuni, C. coli and C. upsaliensis isolated from the stool of human patients in France and Belgium (Vandenberg et al. Citation2004; Prouzet-Mauleon et al. Citation2006; Van den Abeele et al. Citation2014). Houf and Stephan (Citation2007) isolated Arcobacter from seven of 500 (1.4%) stool samples of healthy people with A. cryaerophilus being the only species detected in an Australian adult with intermittent diarrhea and abdominal pain (Tee et al. Citation1988). Although the role of A. skirrowii in human disease is not yet clear in 2004, the first report of A. skirrowii isolation from an elderly person with chronic diarrhea has been published (Wybo et al. Citation2004).
Cases of bacteremia common for both A. butzleri and A. cryaerophilus have infrequently been documented (On et al. Citation1995; Hsueh et al. Citation1997; Yan et al. Citation2000; Lau et al. Citation2002). Recently, a case of A. butzleri bacteremia was reported from a patient with chronic lymphocytic leukemia (Arguello et al. Citation2015). A. butzleri was isolated from blood of a 69-year-old woman (Lau et al. Citation2002). Woo et al. (Citation2001) reported isolation of A. cryaerophilus from a blood sample of a boy who developed acute respiratory distress and succumbed. Also in a case of a newborn bacteremia wherein vertical/transplacental transmission was assumed as route of transmission (On et al. Citation1995).
Reports of Arcobacter occurrence were few from North America earlier, but the current status shows that the pathogen has been reported from Argentina, Brazil, Chile, Costa Rica and also from Mexico (Arias et al. Citation2011). The use of various molecular tools in Portugal has revealed that Arcobacter was prevalent in pediatric people (Ferreira et al. Citation2014c). A. butzleri induced more secretion of pro-inflammatory cytokines, namely TNF, IFN-γ, IL-6 and MCP-1 in small and large intestines. A. butzleri also induces more pronounced local and systemic immune responses than commensal E. coli in a strain-dependent manner. These data suggest that A. butzleri is more than a commensal in vertebrate hosts (Gölz et al. Citation2016a). Zur Bruegge et al. (Citation2016) studied the role of miRNAs in the immune signaling of macrophages infected with A. butzleri. Reports showed several miRNA expressions during bacterial infection but authors also found the up-regulated expression of novel miRNAs like miR-155, miR-212 and miR-125 which has a key role in Toll-like receptor signaling (involved in production of immune response). This study reported the interaction of human innate immune cells with Arcobacter during the course of infection..
10. Diagnosis
Identification of Arcobacter by different biochemical tests is difficult as these organisms are metabolically inert (Collado & Figueras Citation2011), thus isolation of these organisms remains gold standard method for reaching to diagnostic conclusion. Due to lack of accurate detection methods there is no clear data regarding the exact prevalence of Arcobacter from different parts of the world (Fallas-Padilla et al. Citation2014). Development of efficient diagnostic methods could lead to more accurate reporting of human infections and the exact prevalence rate of Arcobacters (Prouzet-Mauleon et al. Citation2006; Merga et al. Citation2011). Current advances in the area of developing rapid and confirmatory diagnostics have provided various molecular methods for rapid and specific detection and differentiation of the Arcobacters at genus and species level with higher sensitivity and specificity.
10.1. Media for isolation of Arcobacters
Arcobacters were first recovered and isolated from bovine and porcine fetal tissues cultured in semi-solid Ellinghausen—McCullough–Johnson–Harris (EMJH) medium supplemented with 5-fluorouracil (Ellis et al. Citation1977; Merga et al. Citation2011). Johnson and Murano (Citation1999a) developed Johnson–Murano (JM) broth and plates, and Houf et al. (Citation2001) developed an Arcobacter-specific isolation method involving the use of Arcobacter medium (Oxoid, United Kingdom) with addition of five antimicrobials (cefoperazone, trimethoprim, amphotericin, novobiocin and 5-fluorouracil). Other isolation methods included EMJH p-80 and Brucella broth (Ongor et al. Citation2004) and direct filtration onto agar without antibiotics via a membrane (Merga et al. Citation2011). Houf et al. (Citation2001a) proposed media with cefoperazone, amphotericin B and teicoplanin (CAT) supplement and EMJH P80 with 5-fluorouracil were sufficient to isolate three Arcobacter species, namely A. butzleri, A. cryaerophilus and A. skirrowii. Arcobacter broth supplemented with CAT was also evaluated for the recovery of these three species (Atabay & Corry Citation1998; Rahimi Citation2014). With 5%–7% defibrinised blood, the samples were incubated on to the Arcobacter media for 48 h at 30 °C under microaerophilic conditions (Aydin et al. Citation2007; Ramees et al. Citation2014b).
Modified charcoal cefoperazone deoxycholate (mCCDA) agar supplemented with CAT antibiotics was used for the isolation of Arcobacter from poultry carcasses (Lammerding et al. Citation1994, Citation1996). de Boer et al. (Citation1996) developed an Arcobacter selective enrichment broth (ASB) and Arcobacter selective semisolid medium (ASM) for the recovery of Arcobacter from food. Collins et al. (Citation1996) developed modified cefsulodin-irgasan-novobiocin (mCIN) agar with CAT supplement for the recovery of Arcobacter from poultry. Atabay et al. (Citation1996) tested the suitability of Karmali medium, mCCDA agar, semi-solid blood-free selectivity-motility (SSM) medium and CAT medium to grow Arcobacters and Campylobacters. Two strains of A. butzleri grew well only on Karmali medium and SSM. A. skirrowii grew poorly on all the selective media used, while A. cryaerophilus did not grow on any of the selective media used. Atabay and Corry (Citation1997) compared CAT agar and mCCDA for the growth of various Arcobacter strains and found that CAT agar performed better. Merga et al. (Citation2011) compared different Arcobacter spp. isolation methods from feces like Houf broth and Houf plates (HH), Houf broth with mCCDA-CAT plates (HCC), Arcobacter broth-CAT broth with Houf plates (ACH), Arcobacter broth-CAT broth with mCCDA-CAT plates (ACCC) and Campylobacter-specific broth and Campylobacter-specific plates (CC). This study revealed that HCC method had higher sensitivity (71%) and specificity (64%).
Johnson and Murano (Citation1999a, Citation1999b) developed an enrichment broth and agar medium to eliminate the growth of Campylobacter species. A. butzleri, A. cryaerophilus and A. nitrofigilis produced red colonies on the medium. Engberg et al. (Citation2000) and Atabay et al. (Citation2002) found that the membrane filtration method is superior to the isolation method without filtration as the percentage of positive samples increased after filtration. Van Driessche et al. (Citation2003) proposed Arcobacter selective isolation broth (ASIB) and Arcobacter selective isolation agar (ASIA) for selective isolation of Arcobacter from feces.
Hamill et al. (Citation2008) compared different media for the isolation of Arcobacter spp. from retail packs of beef and reported that the modified enrichment and selective media of Johnson and Murano gave the highest recovery (15%) of Arcobacters. However, direct detection from the CAT broth by m-PCR and identification of isolates recovered by culture after passive filtration of the broth on blood agar (without any antibiotic supplement) produced more or less the same results in other studies (Collado et al. Citation2008, Citation2009a, Citation2010). A new culture approach was recently adopted for enhanced isolation of Arcobacter from marine samples. Addition of NaCl to the growth media enhances the number of positive samples (Salas-Massó et al. Citation2016). Possibility of isolation of A. butzleri increased when the samples are pre-enriched before plating (Levican et al. Citation2016).
10.2. Detection of Arcobacter spp. by molecular methods
Cultural differentiation is very difficult between Arcobacter and Campylobacter because of their phenotypic similarity and so molecular methods are more useful (Douidah et al. Citation2010; Collado & Figueras Citation2011). Similarly, PCR combined with cultural methods has shown good results in drafting the picture of Arcobacter from sea water in Italy (Maugeri et al. Citation2005). This has led to the development of many different molecular detection methods like PCR, multiplex PCR (m-PCR) and real-time PCR, restriction fragment length polymorphism (RFLP), matrix-assisted laser desorption ionization mass spectrometry (MALDITOF MS), denaturing gradient gel electrophoresis PCR (DGGE-PCR) and fluorescence in situ hybridization (FISH) (Petersen et al. Citation2007; Alispahic et al. Citation2010; Collado & Figueras Citation2011; Patyal et al. Citation2011; Mohan et al. Citation2014).
Detection of Arcobacter in chicken meat at genus level by PCR was developed by González et al. (Citation2014) which has been suggested to be a sensitive tool for epidemiological survey. A nested PCR targeting the 16S rRNA gene to detect Arcobacter spp. and Helicobacter spp. was developed by Suarez et al. (Citation1997). Multiplex PCR to detect both C. jejuni and A. butzleri in the food products was proposed by Winters and Slavik (Citation2000). González et al. (Citation2000) developed combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat following a short selective enrichment of samples. Multiplex real-time PCR was developed for detection of A. butzleri and several Campylobacter spp. from stool samples (de Boer et al. Citation2013). Neubauer and Hess (Citation2006) developed a multiplex PCR protocol which can detect the three closely related genera, i.e. Campylobacter, Helicobacter and Arcobacter in a single reaction cycle from poultry products using primers targeted to amplify the 16S rRNA genes. Wesley et al. (Citation1995) have designed Arcobacter genus and species specific 16S and 23S rRNA probes. Bastyns et al. (Citation1995) developed a genus-specific PCR assay for Arcobacter strains using the variable regions of 23S rRNA. A multiplex PCR assay was developed by Harmon and Wesley (Citation1997) to identify Arcobacter isolates, in particular A. butzleri. Harmon and Wesley (Citation1996) developed a PCR assay targeting 16S rRNA for the identification of A. butzleri, A. cryaerophilus and A. skirrowii. Using a variable 16S rRNA and 23S rRNA region, Houf et al. (Citation2000) developed a species-specific multiplex PCR assay for the simultaneous detection and identification of A. butzleri, A. cryaerophilus and A. skirrowii. Kabeya et al. (Citation2003b) developed an Arcobacter species-specific PCR assay for the detection of A. butzleri, A. cryaerophilus 1A and 1B and A. skirrowii. Levican and Figueras (Citation2013) compared and also reviewed commonly used molecular methods for detection of Arcobacter spp. Authors reported that mPCR of Houf et al. (Citation2000) was most commonly referred followed by Figueras et al. (Citation2008) which employed 16S rDNA-RFLP and then m-PCR of Douidah et al. (Citation2010). However, Levican and Figueras (Citation2013) found that none of the five PCR developed by different authors (Houf et al. Citation2000; Kabeyaet al. Citation2003a; Figueras et al. Citation2008; Pentimalli et al. Citation2009; Douidah et al. Citation2010) were reliable. Authors concluded that result was bad for identification of A. cryaerophilus and A. butzleri using 23S rRNA gene as the target (Levican & Figueras Citation2013). Samarajeewa et al. (Citation2015) compared four diagnostic assays namely next-generation sequencing (Ion torrent technology), DGGE, clonal restriction fragment length polymorphism (C/RFLP) and cloning-sequencing methods for detection of microbes and concluded that all the methods detected Arcobacter spp. Sequencing of PCR products and use of chaperonin sequence database (cpnDB) can help in differentiation of Campylobacter, Arcobacter and Helicobacter isolates (Hill et al. Citation2006).
PCR plus ELISA (Antolin et al. Citation2001) and real-time PCR assays (qRT-PCR) (Abdelbaqi et al. Citation2007a; Brightwell et al. Citation2007) have also been developed for detection of Arcobacter. A SYBR green real-time PCR assay was developed for rapid detection of Arcobacter in food and wastewater samples at the genus level (González et al. Citation2010). PCR-RFLP was developed for identification of three different pathogens namely Campylobacter, Arcobacter and Helicobacter (Marshall et al. Citation1999). Vergis et al. (Citation2013) developed a 16s rRNA PCR combined with RE digestion using EcoRI, HindIII and SalI enzymes which differentiated eight food-borne pathogens (Listeria, Escherichia, Salmonella, Shigella, Vibrio, Campylobacter, Yesinia and Arcobacter). A 16S rDNA-RFLP method was developed using MseI enzyme for six important species of Arcobacter including two hybridization groups of A. cryaerophilus (Figueras et al. Citation2008). Later as the number of species in the genus increased to 17 as per the record of 2012, RFLP studies using the same enzyme MseI yielded distinct pattern for 10 species (A. nitrofigilis, A. cryaerophilus, A. skirrowii, A. cibarius, A. halophilus, A. mytili, A. molluscorum, A. ellisii, A. bivalviorum and A. cloacae) while other species namely A. marinus, A. venerupis, A. defluvii, A. suis, A. butzleri, A. thereius and A. trophiarum showed similar pattern. Hence, different enzymes namely MnlI and BfaI were used to differentiate these species. This study also documented the microheterogenicity among Arcobacter spp. (Figueras et al. Citation2012).
Pentimalli et al. (Citation2009) developed a PCR assay using species-specific primers for the detection of A. butzleri, A. cryaerophilus, A. skirrowii and A. cibarius in chicken meat. A multiplex-PCR assay with seven primers was developed by Douidah et al. (Citation2010) for the identification of the five human and mammal related species of Arcobacter (A. butzleri, A. cryaerophilus, A. skirrowii, A. thereius and A. cibarius). Recently, Webb et al. (Citation2016) developed the PCR primers for direct detection and quantification of A. butzleri DNA in microbiologically complex matrices. Densities of A. butzleri DNA in diarrheic stools were higher when compared to non-diarrheic stools in southwestern Alberta, Canada. The study also reported that among 892 A. butzleri positive diarrheic samples, 74% were not positive for other bacterial or viral pathogens. Miller et al. (Citation2014) formulated a new method for identification of Arcobacter and Campylobacter by using single primer pair to amplify the region of atpA gene followed by sequencing. MALDI-TOF MS has been developed for fast and accurate identification of Arcobacter, Helicobacter and Campylobacter species approving to be a reliable assay (Alispahic et al. Citation2010). Whiteduck-Léveillée et al. (Citation2016a) developed and evaluated the multiplex PCR technique for quick detection of VAGs in Arcobacter spp. This technique could be useful for the detection and prevalence of VAGs in the Arcobacter spp.
Recently developed diagnostic techniques at field level like loop-mediated isothermal amplification (LAMP) test is very useful in pathogen detection with speed and accuracy (Karthik et al. Citation2014a). LAMP has been designed for various pathogens and has also been developed for detection of Arcobacter spp. (Dhama et al. Citation2014a; Karthik et al. Citation2014b; Wang et al. Citation2014). Results showed that the developed LAMP assay was more sensitive than the other molecular assays like multiplex PCR and conventional PCR that were compared in the study (Wang et al. Citation2014). Chicken meat samples were used in the study and LAMP assay could detect Arcobacter spp. specifically and sensitively. The major issue with molecular methods for detection of pathogen is that these assays cannot guarantee the viability of the organisms hence status of infection cannot be assessed clearly (Samarajeewa et al. Citation2015).
An overview of different molecular diagnostic assays for detection of Arcobacters along their advantages and disadvantages/limitations is presented in .
Table 3. An overview of molecular diagnostic assays for detection of Arcobacters.
10.3. Genotyping and determining genetic diversity of Arcobacter spp.
In epidemiological surveillance, typing plays a crucial role to study risk factors and routes of transmission. Several techniques have been tried for strain differentiation of Arcobacters and to trace the source of infection or routes of transmission. These include the use of different PCR-based methods like ERIC-PCR which is most commonly used to trace the different outbreaks (Vandamme et al. Citation1993), to characterize different isolates (Aydin et al. Citation2007) and to study genetic diversity among them (Houf et al. Citation2002b; Collado et al. Citation2010; Suelam Citation2012). Other methods include RAPD-PCR, AFLP and PFGE (Houf et al. Citation2002b; On et al. Citation2002; Ongor et al. Citation2004). ERIC-PCR and RAPD-PCR are both valuable techniques for characterizing A. butzleri, A. cryaerophilus and A. skirrowii isolates. Both methods have satisfactory type ability and discriminatory power. The fingerprints generated with ERIC-PCR were more reproducible and complex than the fingerprints generated with RAPD-PCR (Houf et al. Citation2002b). Other recently developed nucleic acid approaches increasingly being considered for detection, identification and monitoring of Arcobacter in foods include PCR plus RFLP (Neubauer et al. Citation2003), PCR plus RAPD (Atabay et al. Citation2002; Houf et al. Citation2002b) and PCR plus DNA sequencing (Karenlampi et al. Citation2004).
Quinones et al. (Citation2007) used DNA oligonucleotide arrays to simultaneously determine the presence of Arcobacter and Campylobacter in retail chicken samples by targeting VAGs. Arcobacter having wide genetic diversity among the species and between the species have been isolated from different sources (Houf et al. Citation2002b; Merga et al. Citation2013; Gilbert et al. Citation2014). Several researchers stated that there is a high level of diversity among A. butzleri isolates obtained from both human and animal sources as detected by micro restriction PFGE (Hume et al. 2001; Rivas et al Citation2004; González et al. Citation2007a; Merga et al. Citation2013) and ERIC–PCR (Houf et al. Citation2002b; Van Driessche et al. Citation2005, Van Driessche & Houf Citation2007a; De Smet et al. Citation2010). PFGE using restriction enzyme digestion with KpnI has been suggested as the primary method for typing the isolate but huge isolates cannot be typed using this technique. Douidah et al. (Citation2014b) proposed for the use of ERIC-PCR for typing large number of isolates instead of PFGE analysis with KpnI. Son et al. (Citation2006) studied the diversity of Arcobacter and Campylobacter spp. during the processing of broiler carcasses and found that Arcobacter was more diverse genetically than Campylobacter spp. Previous reports suggest that different genotypes have been found in the feces of a single animal (Van Driessche et al. Citation2004, Citation2005). In a case from a pig, 7 A. butzleri, 10 A. cryaerophilus and 6 A. skirrowii genotypes were documented, whereas 6 A. cryaerophilus and 2 A. skirrowii genotypes were reported from cows (Van Driessche et al. Citation2004, Citation2005). First MLST scheme and the first website database of MLST (http://pubmlst.org/arcobacter/) for Arcobacter spp. was done by Miller et al. (Citation2009), set of 374 isolates of Arcobacter (275 A. butzleri, 72 A. cryaerophilus, 15 A. skirrowii and 8 A. cibarius) were screened and concluded that there is no correlation of MLST alleles with host or geographical source. Arcobacter MultiLocus Sequence Typing website (http://pubmlst.org/arcobacter/) provides valuable information for primers and sequencing conditions for the seven genes (atpA, aspA, gltA, glnA, pgm, tkt and glyA) and for submitting new sequences (Miller et al. Citation2009). MLST study using 45 isolates from food products like 13 from raw milk, 12 isolates from poultry, 10 from clams, 5 from pork meats, 4 from mussels and 1 from minced beef in Northern Spain revealed great diversity among A. butzleri isolates and also reported the lateral gene transfer at glyA locus from A. skirrowi to A. butzleri (Alonso et al. Citation2014). Single-enzyme amplified fragment length polymorphism (s-AFLP) technique and PFGE used in tandem could differentiate A. butzleri from A. cryaerophilus and A. skirrowii isolated from water and chicken (González et al. Citation2007b).
Isolates of Arcobacter spp. from water samples along the Llobregat River catchment, at sewage effluents and in a drinking water treatment plant showed high genotypic diversity as revealed by the ERIC-PCR (Collado et al. Citation2010). The workers here reported that out of 339 investigated isolates, 308 had different genotypes among which 248 belonged to the 275 isolates of A. butzleri and 60 to the 63 isolates of A. cryaerophilus were found genotypically diverse. MLST of 32 isolates from swabs of boiler processing line resulted in 10 new sequence types (STs) (Rasmussen et al. Citation2013). A total of 49 Arcobacter isolates (27 A. butzleri and 22 A. cryaerophilus) recovered from a 506 samples (chicken meat, poultry skin, dairy cow milk and human stool) when tested by ERIC-PCR revealed A. butzleri isolates to be grouped into 18 subtypes and A. cryaerophilus isolates grouped into 14 subtypes (Ramees et al. Citation2014d). Genotyping of A. butzleri from cattle in the United Kingdom using MLST and whole genome sequencing revealed an unexpected amount of diversity (Merga et al. Citation2013).
In another study, isolates of Campylobacter, Arcobacter and Helicobacter from reptiles were compared using AFLP atpA and 16S rRNA sequencing (Gilbert et al. Citation2014). Recently, a rapid, high-throughput, high-resolution assay, comparative genomic fingerprinting analysis, has been developed for genotyping of A. butzleri isolates by the use of which reproducible subtyping of these organisms is possible (Webb et al. Citation2015). Development of rapid diagnostic assays will aid in early diagnosis of this food-borne pathogen so that further transmission of this pathogen into the food chain can be prevented.
Advancements in developing rapid, reliable and confirmatory diagnostics along with adapting effective surveillance and monitoring strategies need to be explored to their full potential for countering Arcobacters and their food-borne zoonotic impacts. For this purpose, further strengthening of the immunodiagnostics, molecular detection and characterization, genotyping and differentiation of Arcobacters is suggested by employing advances in PCR and its allied versions, LAMP, recombinant diagnostics, lateral flow assay, biochips, biosensors, microarrays, gene sequencing and phylogenetic analysis, genomic fingerprinting and nanotechnology-based diagnostics (Schmitt & Henderson Citation2005; Deb & Chakraborty Citation2012; Dhama et al. Citation2012, Citation2014a; Num & Useh Citation2013; Gilbert et al. Citation2014; Ramees et al. Citation2014d; Webb et al. Citation2015, Citation2016; Whiteduck-Léveillée et al. Citation2016a).
11. Antibiotic sensitivity and emerging drug resistance
Regarding antimicrobial susceptibility of Arcobacter, very limited data are available and A. butzleri is the only species which is mostly discussed (Kiehlbauch et al. Citation1992; Atabay et al. Citation2003; Houf et al. Citation2001; Fera et al. Citation2003; Kabeya et al. Citation2004). Available reports suggest that there is an increase in antimicrobial resistance against this emerging food-borne pathogen leading to treatment failures with commonly used antimicrobials. A. butzleri is comparatively more resistant than A. cryophilus and A. skirrowi (Houf et al. Citation2001; Kabeya et al. Citation2004; Abay et al. Citation2012). In a study conducted by Kabeya et al. (Citation2004), all the Arcobacter strains tested were found susceptible to ampicillin. Several other studies indicated that Arcobacter are susceptible to aminoglycosides and tetracycline (Fera et al. Citation2003; Son et al. Citation2007; Collado & Figueras Citation2011; Abay et al. Citation2012). Shah et al. (Citation2013) showed that A. butzleri were resistant to ampicillin (56%), followed by cefotaxime (33%) and ciprofloxacin (33%) and susceptible to enrofloxacin and gentamicin, the drugs of choice for the treatment for Arcobacter infection. But in a study conducted by Unver et al. (Citation2013) all of the A. skirrowii and most of the A. cryaerophilus isolates were found susceptible to amoxicillin/clavulanic acid and resistant towards optochin, vancomycin, fusidic acid, cloxacillin and cefazolin with moderate susceptibility to amikacin, enrofloxacin, ofloxacin, oxytetracycline, chloramphenicol, nitrofurantoin, erythromycin, ampicillin sulbactam and amoxicillin. Resistance pattern against antibiotics like cephalothin, novobiocin and vancomycin has been reported by several workers whereas in most cases it was found sensitive against azithromycin, nalidixic acid and gentamicin (Mohan et al. Citation2014). Antimicrobial susceptibilities of 71 Arcobacter isolates were tested against 14 drugs using the disk diffusion method, all of the Arcobacter isolates tested were found to be resistant to one or more antimicrobial agents. Resistance to cephalothin and vancomycin (96%) was the most common finding, followed by resistance to methicillin, azithromycin and ampicillin. All the Arcobacter isolates were susceptible to gentamicin, streptomycin, tetracyclin and kanamycin (Rahimi et al. 2014).
Van den Abeele et al. (Citation2016) reported the antimicrobial susceptibility of A. butzleri and A. cryaerophilus strains isolated from Belgian patients. Majority of the Arcobacter strains were susceptible to gentamicin (99%) and tetracycline (89%). However, erythromycin (78%), ciprofloxacin (72%) and doxycycline (76%) showed moderate activity against Arcobacter spp. Only 9% of the strains were susceptible to ampicillin. Majority of the A. butzleri strains were susceptible to ciprofloxacin (87%), whereas half of the A. cryaerophilus isolates (51%) showed high-level resistance (MIC >32 mg/L). Ciprofloxacin-resistant strains harbored an identical mutation in gyrA. These results suggest that A. cryaerophilus showed an acquired resistance to fluoroquinolones. Macrolides are not first-choice empirical antibiotics for Arcobacter infections. Tetracyclines can be suggested for treatment of Arcobacter induced gastrointestinal infections. Shirzad Aski et al. (Citation2016) conducted an antimicrobial susceptibility study of Arcobacter spp. isolated from clinically healthy food animals. All the Arcobacter isolates were resistant to vancomycin, rifampicin, trimethoprim, ceftriaxone and cephalothin, whereas isolates were highly susceptible to oxytetracycline, tetracycline, ciprofloxacin, erythromycin, kanamycin, amikacin, enrofloxacin and gentamicin. Tetracycline and aminoglycosides can be used for treatment of human Arcobacter infections. The resistance mechanisms of Arcobacter for antibiotics were of mainly chromosomal in nature (Abdelbaqi et al. Citation2007b; Miller et al. Citation2007; Douidah et al. Citation2014a; Ferreira et al. Citation2016).
12. Prevention and control
For food-borne pathogens the control measures are very important to reduce the disease burden (Dhama et al. Citation2013a). Improperly cooked and contaminated food of animal origin like meat, milk, sea food could act as an important source of Arcobacter infection to humans (Collado et al. Citation2009a; Ertaset al. Citation2010; Iwamoto et al. Citation2010; Shah et al. Citation2011). Therefore, thorough and good cooking practices help in prevention of food-borne infections. A. butzleri is more resistant in food samples than C. jejuni to irradiation or cold pasteurization with average D10 value of 0.27 kGy compared with 0.19 kGy for C. jejuni (Phillips Citation2001). Therefore, radiation doses recommended for pork in United States is 0.3–1.0 kGy. Citric acid and lactic acid can eliminate the growth of Arcobacter. At a concentration of 1.0% and 2.0%, citric and lactic acid, respectively can inhibit the growth of A. butzleri, and 2% sodium lactate also effectively checks Arcobacter (Phillips Citation1999; Lehner et al. Citation2005). In a study conducted with nisin alone or in combination with agents like citric acid, lactic acid, sodium citrate or sodium lactate showed that approximately 50% survivability of Arcobacter can be reduced by the use of nisin alone at 500 IU/ml (Phillips Citation1999). Trisodium phosphate and EDTA have been found to be effective in reducing survival of A. butzleri in pure cultures (Phillips & Duggan Citation2001). Low temperature storage can also reduce Arcobacter growth (Phillips Citation2001; Phillips & Long Citation2001). A study on the effect of 17 organic acids (acetic, propionic, butyric, caproic, caprylic, capric, lauric, myristic, palmitic, phenylace-tic, sorbic, benzoic, fumaric, succinic, lactic, malic and citric acid) on Arcobacter spp. showed, that the maximum inhibitory activities were observed with benzoic, citric, malic and sorbic acids (Skřivanová et al. Citation2011). Arcobacters can effectively colonize the surface of pipes such as copper, stainless steel, and plastics (Assanta et al. Citation2002). But regardless of this bacterium's susceptibility to chlorine (Rice et al. Citation1999; Moreno et al. Citation2004) it is still doubtful that whether conventional treatments at a drinking water treatment plant can effectively remove Arcobacter, as failure of treatment could lead to infection (Ho et al. Citation2006a; Collado et al. Citation2010). Since contaminated water acts as an important source of infection, hence the effective treatment of water resources is imperative (Snelling et al. Citation2006; Collado et al. Citation2010). Meat also acts as an important source of Arcobacter transmission, therefore maintenance of slaughter hygiene is crucial for prevention and control of such meat-borne infection. Appropriate hygiene in the slaughter house and dairy plants combined with use of proper disinfectant is needed to eliminate Arcobacter as they could survive even after use of common sanitization protocol (Giacometti et al. Citation2013; Serraino & Giacometti Citation2014). Adherence to practices like strict microbiological monitoring of carcasses, use of verification systems, HACCP and good manufacturing practices are needed to be executed (Lehner et al. Citation2005).
In the era of emerging and rising antibiotic resistance against several commonly used antibiotics, there is a demanding need for exploring alternative and novel therapeutic options for countering infection with Arcobacters in a better way, as like being explored to tackle other food-borne pathogens (Dhama et al. Citation2013a, Citation2013b; Tiwari et al. Citation2013; Yadav et al. Citation2016). In this direction, there is an essential need to shift the attention towards various therapeutic regimens like bacteriophages, probiotics, avian egg yolk antibodies, cytokines, RNAi technology, quorum sensing inhibitors, herbs, essential oils, plant derivatives and others, which have been recently documented to possess potent applications against various pathogenic bacteria having high food-borne concerns (Dhama et al. Citation2008, Citation2013b, Citation2014b; Sudhakar et al. Citation2013; Tiwari et al. Citation2014, Citation2016; Gopi et al. Citation2014; Khan et al. Citation2016). Bacteriophage therapy has been used successfully for prevention of various food-borne pathogen like Salmonella, Listeria, etc. Hence, this therapy can also be helpful for prevention and control of Arcobacter infection with good perspectives at slaughter houses thereby checking the entry of this organism into the human food chain (Karthik et al. Citation2014c). Experimental trials with medicinal herb extracts and spices with respect to their antimicrobial potential have demonstrated that chamomile, sage cinnamon, rosemary, bearberry extracts, which are conventionally practiced for gastric trouble, hinder the growth and development of Arcobacter (Cervenka et al. Citation2006). Seventeen essential oils (rosemary, garlic, sage, bearberry, allspice, black pepper, cumin, cinnamon, coriander, thyme, clove, fennel, licorice, St. Johne's wart, ginger, chamomile and mint) when tested against Arcobacters namely A. butzleri, A. cryaerophilus and A. skirrowii, showed inhibition of these organisms. Similarly, various plant derivatives (lemon, sweet orange and bergamot oils) were documented to prevent Arcobacterosis (Fisher et al. Citation2007). Usage of rosemary essential oil while cooking of minced beef has been reported to cause substantial reduction of A. butzleri (Irkin et al. Citation2011). Resveratrol plant extract has been found to show good bactericidal effect on A. butzleri and A. cryaerophilus via inhibiting the metabolic activity, development and cellular functions of these bacteria, particularly affects DNA in the Arcobacter thereby blocking its cell division, hence can be used for designing potential strategies to control this food-borne pathogen (Ferreira et al. Citation2014a; Alagawany et al. Citation2015).
13. Conclusion and future perspectives
Arcobacter is an emerging food-borne pathogen causing serious infections in humans and animals. Nonetheless, various reports on infection due to Arcobacters are becoming continuously available from different countries, still there is no clear documentation on virulence genes/factors that are directly involved in the causation of infection and pathogenesis of this pathogen. Studies are warranted on this aspect so that targeting of the virulence genes could help in preventing spread of infection. Complete genome of different species in the genus needs to be analyzed so as to draw a conclusion regarding the virulence factors and also the pathogenesis of Arcobacter. Hence, focus of research should be aimed in the near future for unraveling the unknown aspects of pathology and pathogenesis of these organisms, which would pave the way for developing suitable control strategies. Exact status of the incidence of Arcobacter in wild animals is presently lacking which can show the role played by these animals in transmission. This will also help in exploring the detailed epidemiology and knowing the real magnitude of infection, understanding various modes of transmission and spread of Arcobacters in a better way, tracing the source of origin, carriers, and revealing the evolutionary mechanisms associated with their diversity and continuous emergence in humans and animals. Veterinarians and medical professionals should be aware of the route of transmission of Arcobacters so as to educate the human community regarding hygienic practices while cooking and also in abattoirs which would further eliminate the chances of spread of these organisms.
There is also an increase in the development of antimicrobial resistance among Arcobacters against commonly used antimicrobials which is worrisome from the public health perspective. Increasing drug resistant scenario demands attention of researchers towards finding novel and alternative therapeutic options to prevent and control Arcobacters in an effective way, which would be also beneficial with regards to public health and food safety viewpoints. Use of herbal active principles, essential oils, bacteriophages, probiotics, avian egg antibodies, RNAi technologies and other novel therapeutic avenues can help to prevent the development of antibiotic resistance and also the spread of Arcobacters. Strengthening and exploiting the full potential of advanced molecular diagnostic tools and techniques is needed for early and confirmatory diagnosis, differentiation and genotyping of Arcobacters. Research should also be directed towards the development of specific selective media as isolation is always the gold standard for confirmation of this bacterial infection. Diagnosis of Arcobacter has presently improved with the development of various molecular methods giving prompt and confirmatory diagnosis of the pathogen with high sensitivity and specificity. Nowadays, PCR based methods and their allied versions are being increasingly used due the rapidity and accuracy of the result obtained. Techniques like microarray expression profile and proteomic studies would further elaborate on in-depth understanding of Arcobacter infection.
Acknowledgments
The authors of this manuscript thank and acknowledge the support of their respective institutes/universities and the Indian Council of Agriculture Research, New Delhi, India.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abay S, Kayman T, Hizlisoy H, Aydin F. 2012. In vitro antibacterial susceptibility of Arcobacter butzleri isolated from different sources. J Vet Med Sci. 74:613–616.
- Abdelbaqi K, Buissonniere A, Prouzet-Mauleon V, Gresser J, Wesley I, Megraud F, Menard A. 2007a. Development of a realtime fluorescence resonance energy transfer PCR to detect Arcobacter species. J Clin Microbiol. 45:3015–3021.
- Abdelbaqi K, Me´nard A, Prouzet-Mauleon V, Bringaud F, Lehours P, Mégraud F. 2007b. Nucleotide sequence of the gyrA gene of Arcobacter species and characterization of human ciprofloxacin-resistant clinical isolates. FEMS Immunol Med Microbiol. 49:337–345.
- Açik MN, Yüksel H, Ulucan A, Çetinkaya B. 2016. The first experimental research on the pathogenicity of Arcobacter butzleri in zebrafish. Vet Microbiol. 189:32–38.
- Adam Z, Whiteduck-Leveillee K, Cloutier M, Chen W, Lewis CT, Lévesque CA, Topp E, Lapen DR, Tambong JT, Talbot G, Khan IU. 2014a. Draft genome sequence of Arcobacter cibarius Strain LMG21996T, isolated from Broiler Carcasses. Genome Announc. 2:e00034–14.
- Adam Z, Whiteduck-Leveillee K, Cloutier M, Tambong JT, Chen W, Lewis CT, Lévesque CA, Topp E, Lapen DR, Talbot G, Khan IU. 2014b. Draft genome sequences of three Arcobacter strains of pig and dairy cattle manure origin. Genome Announc. 2:e00377–14.
- Adam Z, Whiteduck-Léveillée K, Cloutier M, Chen W, Lewis CT, Lévesque CA, Topp E, Lapen DR, Tambong JT, Talbot G, Khan IU. 2014c. Draft genome sequences of two Arcobacter strains isolated from human feces. Genome Announc. 2:e00113–14.
- Adesiji YO, Seibu E, Emikpe BO, Moriyonu BT, Oloke JK, Coker AO. 2012. Serum biochemistry and heamatological changes associated with graded doses of experimental Arcobacter infection in rats. West Afr J Med. 31:186–191.
- Adesiji YO. 2010. Faecal shedding of Arcobacter species following experimental infection in rats: public health implications. Cent Eur J Med. 5:470–474.
- Alagawany M, Farag MR, Dhama K, El-Hack MEA, Tiwari R, Alam GM. 2015. Mechanisms and beneficial applications of resveratrol as feed additive in animal and poultry nutrition: a review. Int J Pharmacol. 11:213–221.
- Alispahic M, Hummel K, Jandreski-Cvetkovic D, Nöbauer K, Razzazi-Fazeli E, Hess M, Hess C. 2010. Species-specific identification and differentiation of Arcobacter, Helicobacter and Campylobacter by full-spectral matrix-associated laser desorption/ionization time of flight mass spectrometry analysis. J Med Microbiol. 59:295–301.
- Alonso R, Girbau C, Martinez-Malaxetxebarria I, Fernández-Astorga A. 2014. Multilocus sequence typing reveals genetic diversity of foodborne Arcobacter butzleri isolates in the North of Spain. Int J Food Microbiol. 191:125–128.
- Amare LB, Sahela AA, Zunita Z, Jalila A, Hassan L. 2011. Prevalence of Arcobacter spp. on chicken meat at retail markets and in farm chickens in Selengor, Malaysia. Food Control. 22:732–736.
- Anderson KF, Kiehlbauch JA, Anderson DC. 1993. Arcobacter (Campylobacter) butzleri-associated diarrheal illness in a nonhuman primate population. Infect Immun. 61:2220–2223.
- Ansari MI, Harb M, Jones B, Hong PY. 2015. Molecular-based approaches to characterize coastal microbial community and their potential relation to the trophic state of Red Sea. Sci Rep. 5:1–10.
- Antolin A, González I, Garcia T, Hernandez PE, Martin R. 2001. Arcobacter spp. enumeration in poultry meat using a combined PCR-ELISA assay. Meat Sci. 59:169–174.
- Arguello E, Otto CC, Mead P, Babady NE. 2015. Bacteremia caused by Arcobacter butzleri in an immunocompromised host. J Clin Microbiol. 53:1448–1451.
- Arias ML, Cid A, Fernandéz H. 2011. Arcobacter butzleri: first isolation report from chicken carcasses in Costa Rica. Braz J Microbiol. 42:703–706.
- Assanta MA, Roy D, Lemay MJ, Montpetit D. 2002. Attachment of Arcobacter butzleri, a new waterborne pathogen, to water distribution pipe surfaces. J Food Prot. 65:1240–1247.
- Atabay HI, Aydin F, Houf K, Sahin M, Vandamme P. 2002. The prevalance of Arcobacter spp. on chicken carcasses sold in retail markets in Turkey, and identification of the isolates using SDS-PAGE. Int J Food Microbiol. 81:21–28.
- Atabay HI, Aydin F, Houf K, Sahin M, Vandamme P. 2003. The prevalence of Arcobacter spp. on chicken carcass sold in retail markets in Turkey and identification of the isolates using SDS-PAGE. Int J Food Microbiol. 25:21–28.
- Atabay HI, Aydin F. 2001. Susceptibility of Arcobacter butzleri isolates to 23 antimicrobial agents. Lett Appl Microbiol. 33:430–433.
- Atabay HI, Corry JEL, On SLW. 1998. Diversity and prevalence of Arcobacter spp. in broiler chickens. J Appl Microbiol. 84:1007–1016.
- Atabay HI, Corry JEL, Prost DE. 1996. Comparison of the productivity of a variety of selective media for Campylobacter and Arcobacter species In: Newell DJ, Ketley JM, Felman RA, editors. Campylobacters, Helicobacters and related organisms New York (NY): Plenum Press; p. 19–23.
- Atabay HI, Corry JEL. 1997. The prevalence of Campylobacters and Arcobacters in broiler chickens. J Appl Microbiol. 83:619–626.
- Atabay HI, Corry JEL. 1998. Evaluation of a new Arcobacter enrichment medium and comparison with new media developed for enrichment for Campylobacter spp. Int J Food Microbiol. 41:53–58.
- Atabay HI, Unver A, Sahin M, Otlu S, Elmali M, Yaman H. 2008. Isolation of various Arcobacter species from domestic geese (Anser anser). Vet Microbiol. 128:400–405.
- Atabay HI, Wain M, Madsen M. 2006. Detection and diversity of various Arcobacter species in Danish poultry. Int J Food Microbiol. 9:139–145.
- Aydin F, Gumussoy KS, Atabay HI, Ica T, Abay S. 2007. Prevalence and distribution of Arcobacter species in various sources in Turkey and molecular analysis of isolated strains by ERIC-PCR. J Appl Microbiol. 103:27–35.
- Badilla-Ramírez Y, Fallas-Padilla KL, Fernández-Jaramillo H, Arias-Echandi ML. 2016. Survival capacity of Arcobacter butzleri inoculated in poultry meat at two different refrigeration temperatures. Rev Inst Med Trop Sao Paulo. 58:22.
- Bagalakote PS, Rathore RS, Ramees TP, Mohan HV, Sumankumar M, Agarwal RK, Kumar A, Dhama K. 2014. Molecular characterization of Arcobacter isolates using randomly amplified polymorphic DNA- polymerase chain reaction (RAPD-PCR). Asian J Anim Vet Adv. 9:543–555.
- Bastyns K, Cartuyvels D, Chapelle S, Vandamme P, Goossens H, De Wachter R. 1995. A variable 23S rDNA region is a useful discriminating target for genus-specific and species-specific PCR amplification in Arcobacter species. System Appl Microbiol. 18:353–356.
- Bath GF, Leask R, Pettey KP, Coetzee DJ. 2013. Abortions in sheep associated with Arcobacter skirrowii infection. J South Afr Vet Assoc. 84:4. Available from: https://doi.org/10.4102/jsava.v84i1.952
- Bogantes EV, Fallas-Padilla KL, Rodríguez-Rodríguez CE, Jaramillo HF, Echandi ML.. 2015. Zoonotic species of the genus arcobacter in poultry from different regions of Costa Rica. J Food Prot. 78:808–811.
- Brann OS. 2001. Infectious complications of cirrhosis. Curr Gastroenterol Rep. 3:285–292.
- Brightwell G, Mowat E, Clemens R, Boerema J, Pulford DJ, On SL. 2007. Development of a multiplex and real time PCR assay for the specific detection of Arcobacter butzleri and Arcobacter cryaerophilus. J Microbiol Methods. 68:318–325.
- Bucker R, Troeger H, Kleer J, Fromm M, Schulzke JD. 2009. Arcobacter butzleri induces barrier dysfunction in intestinal HT-29/B6 cells. J Infect Dis. 200:756–764.
- Calvo G, Arias ML, Fernández H. 2013. Arcobacter: a foodborne emerging pathogen. Arch Latinoam Nutr. 63:164–172.
- Cervenka L, Peskova I, Foltynova E, Pejchalova M, Brozkova I, Vytrasova J. 2006. Inhibitory effects of some spice and herb extracts against Arcobacter butzleri, A. cryaerophilus, and A. skirrowii. Curr Microbiol. 53:435–439.
- Cervenka L. 2007. Survival and inactivation of Arcobacter spp., a current status and future prospect. Crit Rev Microbiol. 33:101–108.
- Chavatte N, Baré J, Lambrecht E, Van Damme I, Vaerewijck M, Sabbe K, Houf K. 2014. Co-occurrence of free-living protozoa and foodborne pathogens on dishcloths: implications for food safety. Int J Food Microbiol. 191:89–96.
- Chinivasagam HN, Corney BG, Wright LL, Diallo IS, Blackall PJ. 2007. Detection of Arcobacter spp. in piggery effluent and effluent-irrigated soils in southeast Queensland. J Appl Microbiol. 103:418–426.
- Collado L, Cleenwerck I, Van Trappen S, De Vos P, Figueras MJ. 2009b. Arcobacter mytili sp. nov., an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int J Syst Evol Microbiol. 59:1391–1396.
- Collado L, Figueras MJ. 2011. Taxonomy, epidemiology and clinical relevance of the genus Arcobacter. Clin Microbiol Rev. 24:174–192.
- Collado L, Guarro J, Figueras M. 2009a. Prevalence of Arcobacter in meat and shellfish. J Food Protect. 72:1102–1110.
- Collado L, Inza I, Guarro J, Figueras M J. 2008. Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environ Microbiol. 10:1635–1640.
- Collado L, Jara R, Vásquez N, Telsaint C. 2014. Antimicrobial resistance and virulence genes of Arcobacter isolates recovered from edible bivalve mollusks. Food Control. 46:508–512.
- Collado L, Kasimir G, Perez U, Bosch A, Pinto R, Saucedo G, Huguet JM, Figueras MJ. 2010. Occurrence and diversity of Arcobacter spp. along the Llobregat river catchment, at sewage effluents and in a drinking water treatment plant. Water Res. 44:3696–3702.
- Collado L, Levican A, Perez J, Figueras MJ. 2011. Arcobacter defluvii sp. nov., isolated from sewage samples. Int J Syst Evol Microbiol. 61:2155–2161.
- Collins CI, Wesley IV, Murano EA. 1996. Detection of Arcobacter spp. in ground pork by modified plating methods. J Food Protect. 59:448–452.
- Corry JE, Atabay HI. 2001. Poultry as a source of Campylobacter and related organisms. J Appl Microbiol. 90:96S–114S.
- D'Sa EM, Harrison MA. 2005. Effect of pH, NaCl content, and temperature on growth and survival of Arcobacter spp. J Food Protect. 68:18–25.
- de Boer E, Tilburg JJHC, Woodward DL, Loir H, Johnson WM. 1996. A selective medium for the isolation of Arcobacter from meats. Lett Appl Microbiol. 23:64–66.
- de Boer RF, Ott A, Güren P, van Zanten E, van Belkum A, Kooistra-Smid AM. 2013. Detection of Campylobacter species and Arcobacter butzleri in stool samples by use of real-time multiplex PCR. J Clin Microbiol. 51:253–259.
- de Oliveira SJ, Wesley I, Harmon KM. 1997. Classification of Arcobacter species isolated from aborted pig fetuses and sows with reproductive problems in Brazil. Vet Microbiol. 57:347–354.
- De Smet S, De Zutter L, Houf K. 2011. Small ruminants as carriers of the emerging food-borne pathogen Arcobacter on small and medium farms. Small Rumin Res. 97:124–129.
- De Smet S, De Zutter L, Van Hende J, Houf K. 2010. Arcobacter contamination on pre- and post-chilled bovine carcasses and in minced beef at retail. J Appl Microbiol. 108:299–305.
- Deb R, Chakraborty S. 2012. Trends in veterinary diagnostics. J Vet Sci Technol. 3:1.
- Debruyne L, Gevers D, Vandamme P. 2008. Taxonomy of the family Campylobacteraceae. In: Nachamkin I, Szymanski C, Blaser M, editors. Campylobacter. Vol. 3. Washington (DC): ASM press; p. 3–25.
- Dhama K, Chakraborty S, Mahima, Wani MY, Verma AK, Deb R, Tiwari R, Kapoor S. 2013b. Novel and emerging therapies safeguarding health of humans and their companion animals: a review. Pak J Biol Sci. 16:101–111.
- Dhama K, Karthik K, Chakraborty S, Tiwari R, Kapoor S, Kumar A, Thomas P. 2014a. Loop-mediated isothermal amplification of DNA (LAMP) – a new diagnostic tool lights the world of diagnosis of animal and human pathogens: a review. Pak J Biol Sci. 17:151–166.
- Dhama K, Mahendran M, Tomar S, Chauhan RS. 2008. Beneficial effects of probiotics and prebiotics in livestock and poultry: the current perspectives. Intas Polivet. 9:1–12.
- Dhama K, Rajagunalan S, Chakraborty S, Verma AK, Kumar A, Tiwari R, Kapoor S. 2013a. Food-borne pathogens of animal origin-diagnosis, prevention, control and their zoonotic significance: a review. Pak J Biol Sci. 16:1076–1085.
- Dhama K, Tiwari R, Chakraborty S, Saminathan M, Kumar A, Karthik K, Wani MY, Amarpal, Singh SV, Rahal A. 2014b. Evidence based antibacterial potentials of medicinal plants and herbs countering bacterial pathogens especially in the era of emerging drug resistance: an integrated update. Int J Pharmacol. 10:1–43.
- Dhama K, Wani MY, Tiwari R, Kumar D. 2012. Molecular diagnosis of animal diseases: the current trends and perspectives. Livestock Sphere. 1:6–10.
- Di Francesco A, Delogu M, Giacometti F, Stancampiano L, Grilli E, Guarniero I, Serraino A. 2014. First detection of Arcobacter sp. in Eurasian collared doves (Streptopelia decaocto). Vet Ital. 50:313–315.
- Diéguez AL, Balboa S, Magnesen T, Romalde JL. 2017. Arcobacter lekithochrous sp. nov., a new species isolated from a molluscan hatchery in Norway. Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.001809.
- Donachie SP, Bowman JP, On SLW, Alam M. 2005. Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter. Int J Syst Evol Microbiol. 55:1271–1277.
- Douidah L, De Zutter L, Baré J, Houf K. 2014b. Towards a typing strategy for Arcobacter species isolated from humans and animals and assessment of the in vitro genomic stability. Foodborne Pathog Dis. 11:272–280.
- Douidah L, De Zutter L, Van Nieuwerburgh F, Deforce D, Ingmer H, Vandenberg O, Van den Abeele AM, Houf K. 2014a. Presence and analysis of plasmids in human and animal associated Arcobacter species. PloS One. 9:e85487.
- Douidah L, Zutter LD, Vandamme P, Houf K. 2010. Identification of five human and mammal associated Arcobacter species by a novel multiplex-PCR assay. J Microbiol Methods. 80:281–286.
- Douidah L, de Zutter L, Baré J, De Vos P, Vandamme P, Vandenberg O, Van den Abeele AM, Houf K. 2012. Occurrence of putative virulence genes in Arcobacter species isolated from humans and animals. J Clin Microbiol. 50:735–741.
- Ellis WA, Neill SD, O’ Brien JJ, Ferguson, HW, Hanna J. 1977. Isolation of Spirillum/Vibrio-like organisms from bovine fetuses. Vet Record. 100:451–452.
- Ellis WA, Neill SD, O’ Brien JJ, Hannna J. 1978. Isolation of Spirullum-like organisms from pig fetuses. Vet Record. 102:106.
- Engberg J, On SLW, Harrington CS, Gerner-Smidt P. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J Clin Microbiol. 38:286–291.
- Ertas N, Dogruer Y, Gonulalan Z, Guner A, Ulger I. 2010. Prevalence of Arcobacter species in drinking water, spring water, and raw milk as determined by multiplex PCR. J Food Prot. 73:2099–2102.
- Fallas-Padilla KL, Rodríguez-Rodríguez CE, Fernández Jaramillo H, Arias Echandi ML. 2014. Arcobacter: comparison of isolation methods, diversity, and potential pathogenic factors in commercially retailed chicken breast meat from Costa Rica. J Food Prot. 77:880–884.
- Fera MT, Gugliandolo C, Lentini V, Favaloro A, Bonanno D, La Camera E, Maugeri TL. 2010. Specific detection of Arcobacter spp. in estuarine waters of Southern Italy by PCR and fluorescent in situ hybridization. Lett Appl Microbiol. 50:65–70.
- Fera MT, La Camera E, Carbone H, Malara D, Pennisi MG. 2009. Pet cats as carriers of Arcobacter spp. in Southern Italy. J Appl Microbiol. 106:1661–1666.
- Fera MT, Maugeri TL, Giannone M, Gugliandolo C, La Camera E, Blandino G, Carbone M. 2003. In vitro susceptibility of Arcobacter butzleri and Arcobacter cryaerophilus to different antimicrobial agents. Int J Antimicrob Agents. 21:488–491.
- Ferna´ndez H, Eller G, Paillacar J, Gajardo T, Riquelme A. 1995. Toxigenic and invasive capacities: possible pathogenic mechanisms in Arcobacter cryaerophilus. Mem Inst Oswaldo. 90:633–634.
- Ferreira S, Júlio C, Queiroz JA, Domingues FC, Oleastro M. 2014c. Molecular diagnosis of Arcobacter and Campylobacter in diarrhoeal samples among Portuguese patients. Diagn Microbiol Infect Dis. 78:220–225.
- Ferreira S, Queiroz JA, Oleastro M, Domingues FC. 2014b. Genotypic and phenotypic features of Arcobacter butzleri pathogenicity. Microb Pathog. 76:19–25.
- Ferreira S, Queiroz JA, Oleastro M, Domingues FC. 2016. Insights in the pathogenesis and resistance of Arcobacter: a review. Crit Rev Microbiol. 42:364–383.
- Ferreira S, Silva F, Queiroz JA, Oleastro, M, Domingues FC. 2014a. Resveratrol against Arcobacter butzleri and Arcobacter cryaerophilus: activity and effect on cellular functions. Int J Food Microbiol. 180:62–68.
- Figueras MJ, Collado L, Guarro J. 2008. A new 16S rDNA-RFLP method for the discrimination of the accepted species of Arcobacter. Diag Microbiol Infect Dis. 62:11–15.
- Figueras MJ, Collado L, Levican A, Perez J, Solsona MJ, Yustes C. 2011a. Arcobacter molluscorum sp. nov., a new species isolated from shellfish. Syst Appl Microbiol. 34:105–109.
- Figueras MJ, Levican A, Collado L, Inza MI, Yustes C. 2011b. Arcobacter ellisii sp. nov., isolated from mussels. Syst Appl Microbiol. 34:414–418.
- Figueras MJ, Levican A, Collado L. 2012. Updated 16S rRNA-RFLP method for the identification of all currently characterised Arcobacter spp. BMC Microbiol. 12:292.
- Figueras MJ, Levican A, Pujol I, Ballester F, Rabada, Quilez MJ, Gomez-Bertomeu F. 2014. A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter sp. and a review of the clinical incidence of Arcobacter spp. New Microbes New Infect. 2:31–37.
- Fisher JC, Levican A, Figueras MJ, McLellan SL. 2014. Population dynamics and ecology of Arcobacter in sewage. Front Microbiol. 5:1–9.
- Fisher K, Rowe C, Phillips CA. 2007. The survival of three strains of Arcobacter butzleri in the presence of lemon, orange and bergamot essential oils and their components in vitro and on food. Lett Appl Microbiol. 44:495–499.
- Flanagan RC, Neal-McKinney JM, Dhillon AS, Miller WG, Konkel ME. 2009. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect Immun. 77:2399–2407.
- Fong TT, Mansfield LS, Wilson DL, Schwab DJ, Molly SL, Rose JB. 2007. Massive microbiological ground water contamination associated with a waterborne outbreak in lake Eri South Bass Island, Ohio. Environ Health Perspect. 115:856–864.
- Giacometti F, Lucchi A, Di Francesco A, Delogu M, Grilli E, Guarniero I, Stancampiano L, Manfreda G, Merialdi G, Serraino A. 2015. Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii circulation in a dairy farm and sources of milk contamination. Appl Environ Microbiol. 81:5055–5063.
- Giacometti F, Lucchi A, Manfreda G, Florio D, Zanoni RG, Serraino A. 2013. Occurrence and genetic diversity of Arcobacter butzleri in an artisanal dairy plant in Italy. Appl Environ Microbiol. 79:6665–6669.
- Giacometti F, Serraino A, Pasquali F, De Cesare A, Bonerba E, Rosmini R. 2014. Behavior of Arcobacter butzleri and Arcobacter cryaerophilus in ultrahigh-temperature, pasteurized, and raw cow's milk under different temperature conditions. Foodborne Pathog Dis. 11:15–20.
- Gilbert MJ, Kik M, Timmerman AJ, Severs TT, Kusters JG, Duim B, Wagenaar JA. 2014. Occurrence, diversity, and host association of intestinal Campylobacter, Arcobacter and Helicobacter in Reptiles. PloS One. 9:e101599.
- Gill KP.1983. Aerotolerant campylobacter strain isolated from a bovine preputial sheath washing. Vet Rec. 112:459.
- Golla SC, Murano EA, Johnson LG, Tipton NC, Cureington EA, Savell JW. 2002. Determination of the occurrence of Arcobacter butzleri in beef and dairy cattle from Texas by various isolation methods. J Food Prot. 65:1849–1853.
- González A, Suski J, Ferrus MA. 2010. Rapid and accurate detection of Arcobacter contamination in commercial chicken products and waste water samples by real-time polymerase chain reaction. Foodborne Pathog Dis. 7:327–338.
- González AM, Ferrus MA. 2011. Study of Arcobacter spp. contamination in fresh lettuces detected by different cultural and molecular methods. Int J Food Microbiol. 145:311–314.
- González I, Garcia T, Antonlin A, Hernandez PE, Martin R. 2000. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat. Lett Appl Microbiol. 30:207–212.
- González A, Botella S, Montes RM, Moreno Y, Ferrus MA. 2007a. Direct detection and identification of Arcobacter species by multiplex PCR in chicken and waste water samples from Spain. J Food Prot. 70:341–347.
- González A, Moreno Y, González R, Herna´ndez J, Ferrus MA. 2006. Development of a simple and rapid method based on polymerase chain reaction-based restriction fragment length polymorphism analysis to differentiate Helicobacter, Campylobacter and Arcobacter species. Curr Microbiol. 53:416–421.
- González A, Ferrús MA, González R, Hernández J. 2007b. Molecular fingerprinting of Campylobacter and Arcobacter isolated from chicken and water. Int Microbiol. 10:85–90.
- González I, Fernández-Tomé S, García T, Martín R. 2014. Genus-specific PCR assay for screening Arcobacter spp. in chicken meat. J Sci Food Agric. 94:1218–1224.
- Gopi M, Karthik K, Manjunathachar HV, Tamilmahan P, Kesavan M, Dashprakash M, Balaraju BL, Purushothaman MR. 2014. Essential oils as a feed additive in poultry nutrition. Adv Anim Vet Sci. 2:1–7.
- Gölz G, Alter T, Bereswill S, Heimesaat MM. 2016a. The Immunopathogenic Potential of Arcobacter butzleri – lessons from a meta-analysis of murine infection studies. PLoS One. 11:e0159685.
- Gölz G, Alter T, Bereswill S, Heimesaat MM. 2016b. Toll-like receptor-4 dependent intestinal gene expression during Arcobacter butzleri infection of gnotobiotic Il-10 deficient mice. Eur J Microbiol Immunol. 6:67–80.
- Gölz G, Karadas G, Alutis ME, Fischer A, Kuhl AA, Breithaupt A, Gobel UB, Alter T, Bereswill S, Heimesaat MM. 2015a. Arcobacter butzleri induce colonic, extra-intestinal and systemic inflammatory responses in gnotobiotic IL-10 deficient mice in a strain-dependent manner. PloS One. 10:e0139402.
- Gölz G, Karadas G, Fischer A, Gobel UB, Alter T, Bereswill S, Heimesaat MM. 2015b. Toll-like receptor-4 is essential for Arcobacter butzleri-induced colonic and systemic immune responses in gnotobiotic IL-10–/–mice. Eur J Microbiol Immunol. 5:321–332.
- Gugliandolo C, Irrera GP, Lentini V, Maugeri TL. 2008. Pathogenic Vibrio, Aeromonas and Arcobacter spp. associated with copepods in the straits of Messina (Italy). Mar Pollut Bull. 56:600–606.
- Hamill S, Neill SD, Madden RH. 2008. A comparison of media for the isolation of Arcobacter spp. from retail packs of beef. J Food Protect. 71:850–854.
- Hamir AN, Sonn RJ, Franklin S, Wesley IV. 2004. Campylobacter jejuni and Arcobacter species associated with intussusceptions in a raccoon (Procyon lotor). Vet Record. 155:338–340.
- Hänel I, Tomaso H, Neubauer H. 2016. Arcobacter - an underestimated zoonotic pathogen? Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 59:789–794.
- Harmon KM, Wesley IV. 1996. Identification of Arcobacter isolates by PCR. Lett Appl Microbiol. 23:241–244.
- Harmon KM, Wesley IV. 1997. Multiplex PCR for the identification of Arcobacter and differentiation of Arcobacter butzleri from other Arcobacters. Vet Microbiol. 58:215–227.
- Harrab B, Schwarz S, Wenzel S. 1998. Identi¢cation and characterization of Arcobacter isolates from broilers by biochemical tests, antimicrobial resistance patterns and plasmid analysis. J Vet Med. 45:87–94.
- Hausdorf L, Fröhling A, Schlüter O, Klocke M. 2011. Analyses of the bacterial community within carrot wash water. Can J Microbiol. 57:447–452.
- Hausdorf L, Neumann M, Bergmann I, Sobiella K, Mundt K, Fröhling A, Schlüter O, Klocke M. 2013. Occurrence and genetic diversity of Arcobacter spp. in a spinach-processing plant and evaluation of two Arcobacter-specific quantitative PCR assay. Syst Appl Microbiol. 36:235–243.
- Heimesaat MM, Alter T, Bereswill S, Gölz G. 2016. Intestinal expression of genes encoding inflammatory mediators and gelatinases during Arcobacter butzleri infection of gnotobiotic Il-10 deficient mice. Eur J Microbiol Immunol. 6:56–66.
- Heimesaat MM, Karadas G, Alutis M, Fischer A, Kuhl AA, Breithaupt A, Gobel UB, Alter T, Bereswill S, Gölz G. 2015a. Survey of small intestinal and systemic immune responses following murine Arcobacter butzleri infection. Gut Pathog. 7:28.
- Heimesaat MM, Karadas G, Fischer A, Gobel UB, Alter T, Bereswill S, Gölz G. 2015b. Toll-like receptor-4 dependent small intestinal immune responses following murine Arcobacter butzleri infection. Eur J Microbiol Immunol. 5:333–342.
- Higgins R, Messier S, Daignault D, Lorange M. 1999. Arcobacter butzleri isolated from a diarrhoeic non-human primate. Lab Anim. 33:87–90.
- Hill JE, Paccagnella A, Law K, Melito PL, Woodward DL, Price L, Leung AH, Ng LK, Hemmingsen SM, Goh SH. 2006. Identification of Campylobacter spp. and discrimination from Helicobacter and Arcobacter spp. by direct sequencing of PCR-amplified cpn60 sequences and comparison to cpnDB, a chaperonin reference sequence database. J Med Microbiol. 55:393–399.
- Hilton CL, Mackey BM, Hargreaves AJ, Forsythe SJ. 2001. The recovery of Arcobacteri NCTC 12481 from various temperature treatments. J Appl Microbiol. 91:929–932.
- Ho HT, Lipman LJ, Gaastra W. 2006a. Arcobacter, what is known and unknown about a potential food-borne zoonotic agent.Vet Microbiol. 115:1–13.
- Ho HT, Lipman LJ, Gaastra W. 2008a. The introduction of Arcobacter spp. in poultry slaughterhouses. Int J Food Microbiol. 125:223–229.
- Ho HT, Lipman LJ, Hendriks HG, Tooten PC, Ultee T, Gaastra W. 2007. Interaction of Arcobacter spp. with human and porcine intestinal epithelial cells. FEMS Immunol Med Microbiol. 50:51–58.
- Ho HT, Lipman LJ, Wosten MM, van Asten AJ, Gaastra W. 2008b. Arcobacter spp. possess two very short flagellins of which FlaA is essential for motility. FEMS Immunol Med Microbiol. 53:85–95.
- Ho HT, Lipman LJA, Van der Graaf-van Bloois L, Van Bergen M, Gaastra W. 2006b. Potential routes of acquisition of Arcobacter species by piglets. Vet Microbiol. 114:122–133.
- Houf K, De Zutter L, Van Hoof J, Vandamme P. 2002a. Occurrence and distribution of Arcobacter species in poultry processing. J Food Prot. 65:1233–1239.
- Houf K, De Zutter L, Van Hoof J, Vandamme P. 2002b. Assessment of the genetic diversity among arcobacters isolated from poultry products by using two PCR-based typing methods. Appl Environ Microbiol. 68:2172–2178.
- Houf K, Devriese LA, De Zutter L, Verbeke B, Van Hoof J, Vandamme P. 2003. Molecular characterization of Arcobacter isolates collected in a poultry slaughterhouse. J Food Prot. 66:364–369.
- Houf K, Devriese LA, De zutter L, Van Hoof J, Vandamme P. 2001. Susceptibility of Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii to antimicrobial agents used in selective media. J Clin Microbiol. 39:1654–1656.
- Houf K, On SL, Coenye T, Debruyne L, De Smet S, Vandamme P. 2009. Arcobacter thereius sp. nov., isolated from pigs and ducks. Int J Syst Evol Microbiol. 59:2599–2604.
- Houf K, Sarah DS, Julie B, Sylvie D. 2008. Dogs as carriers of the emerging pathogen; Arcobacter. J Vet Microbiol. 130:208–213.
- Houf K, Stephan R. 2007. Isolation and characterization of the emerging food-borne pathogen Arcobacter from human stool. J Microbiol Methods. 68:408–413.
- Houf K, Tutenel A, De Zutter L, Van Hoof J, Vandamme P. 2000. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Immunol Med Microbiol. 49:337–345.
- Hsu TT, Lee J. 2015. Global distribution and prevalence of arcobacter in food and water. Zoonoses Public Health. 62:579–589.
- Hsueh PR, Teng LJ, Yang, PC, Wang SK, Chang SC, Ho SW, Hsieh WC, Luh KT. 1997. Bacteremia caused by Arcobacter cryaerophilus 1B. J Clin Microbiol. 35:489–491.
- Hu Q, Lyu D, Shi X, Jiang Y, Lin Y, Li Y, Qiu Y, He L, Zhang R, Li Q. 2014. A modified molecular beacons-based multiplex real-time pcr assay for simultaneous detection of eight foodborne pathogens in a single reaction and its application. Foodborne Pathog Dis. 11:207–214.
- Irkin R, Abay S, Aydin F. 2011. Inhibitory effects of some plant essential oils against Arcobacter butzleri and potential for rosemary oil as a natural food preservative. J Med Food. 14:291–296.
- Iwamoto M, Ayers T, Mahon BE, Swerdlow DL. 2010. Epidemiology of seafood-associated infections in the United States. Clin Microbiol Rev. 23:399–411.
- Jacob J, Loir H, Feuerpfeil I. 1993. Isolation of Arcobacter butzleri from drinking water reservoir in Eastern Germany. Int J Hyg Env Med. 193:557–562.
- Jacob J, Woodward D, Feuerpfeil I, Johnson WM. 1998. Isolation of Arcobacter butzleri in raw water and drinking water treatment plants in Germany. Zbl Hyg. 201:189–198.
- Jiang ZD, DuPont HL, Brown EL, Nandy RK, Ramamurthy T, Sinha A, Ghosh, S, Guin S, Gurleen K, Rodrigues S, et al. 2010. Microbial etiology of travelers’ diarrhea in Mexico, Guatemala, and India: importance of enterotoxigenic Bacteroides fragilis and Arcobacter species. J Clin Microbiol. 48:1417–1419.
- Johnson LG, Murano EA. 1999a. Development of a new medium for the isolation of Arcobacter spp. J Food Prot. 62:456–462.
- Johnson LG, Murano EA. 1999b. Comparison of three protocols for the isolation of Arcobacter from poultry. J Food Prot. 62:610–614.
- Kabeya H, Koboyashi Y, Maruyama S, Mikami T. 2003b. One step polymerase chain reaction-based typing of Arcobacter species. Int J Food Microbiol. 81:163–168.
- Kabeya H, Maruyama S, Morita Y, Kubo M, Yamamoto K, Arai S, Izumi T, Kobayashi Y, Katsube Y, Mikami T. 2003a. Distribution of Arcobacter species among livestock in Japan. Vet Microbiol. 93:153–158.
- Kabeya H, Maruyama S, Morita Y, Ohsuga T, Ozawa S, Kobayashi Y, Abe M, Katsube Y Mikami T. 2004. Prevalence of Arcobacter species in retail meats and antimicrobial susceptibility of the isolates in Japan. Int J Food Microbiol. 90:303–308.
- Karadas G, Sharbati S, Hänel I, Messelhäußer U, Glocker E, Alter T, Gölz G. 2013. Presence of virulence genes, adhesion and invasion of Arcobacter butzleri. J Appl Microbiol. 115:583–590.
- Kärenlampi RI, Tolvanen TP, Hänninen ML. (2004). Phylogenetic analysis and PCR-restriction fragment length polymorphism identification of campylobacter species based on partial groEL gene sequences. J Clin Microbiol. 42:5731–5738.
- Karthik K, Muneeswaran NS, Manjunathachar HV, Gopi M, Elamurugan A, Kalaiyarasu S. 2014c. Bacteriophages: effective alternative to antibiotics. Adv Anim Vet Sci. 2:1–7.
- Karthik K, Rathore R, Thomas P, Arun TR, Viswas KN, Agarwal RK, Manjunathachar HV, Dhama K. 2014a. Loop-mediated isothermal amplification (LAMP) test for specific and rapid detection of Brucella abortus in cattle. Vet Quart. 34:174–179.
- Karthik K, Rathore R, Thomas P, Arun TR, Viswas KN, Dhama K, Agarwal RK. 2014b. New closed tube loop mediated isothermal amplification assay for prevention of product cross contamination. MethodsX. 1:e137–e143.
- Khan RU, Naz S, Dhama K, Karthik K, Tiwari R, Abdelrahman MM, Alhidary IA, Zahoor A. 2016. Direct-fed microbial: beneficial applications, modes of action and prospecas safe tool for enhancing ruminant production and safeguarding health. Int J Pharmacol. 12:220–231.
- Khoshbakht R, Tabatabaei M, Shirzad Aski H, Seifi S. 2014. Occurrence of Arcobacter in Iranian poultry and slaughterhouse samples implicates contamination by processing equipment and procedures. Br Poult Sci. 55:732–736.
- Kiehlbauch JA, Baker C, Wachsmuth IK. 1992. In vitro susceptibilities of aerotolerant Campylobacter isolates to 22 antimicrobial agents. Antimicrob Agents Chemother. 36:717–722.
- Kiehlbauch JA, Brenner DJ, Nicholson MA, Baker CN, Patton CM, Steigerwalt AG, Wachsmuth IK. 1991. Campylobacter butzleri sp. nov. isolated from humans and animals with diarrheal illness. J Clin Microbiol. 29:376–385.
- Kim HM, Hwang CY, Cho BC. 2010. Arcobacter marinus sp. nov. Int J Syst Evol Microbiol. 60:531–536.
- Kjeldgaard J, Jorgensen K, Ingmer H. 2009. Growth and survival at chiller temperature of Arcobacter butzleri. Int J Food Microbiol. 131:256–259.
- Kolling G, Wu M, Guerrant RL. 2012. Enteric pathogens through life stages. Front Cell Infect Microbiol. 2:114.
- Kopilovic B, Ucakar V, Koren N, Krek M, Kraigher A. 2008. Waterborne outbreak of acute gastroentritis in a coastal area in Slovenia in June and July 2008. Eurosurveillance. 13:1–3.
- Laishram M, Rathlavath S, Lekshmi M, Kumar S, Nayak BB. 2016. Isolation and characterization of Arcobacter spp. from fresh seafood and the aquatic environment. Int J Food Microbiol. 232:87–89.
- Lammerding AM, Harris JE, Lior H, Woodward D, Cole L, Muckle CA. 1996. Isolation method for recovery of Arcobacter butzleri from fresh poultry and poultry products. In Newell DG, Ketley J, Feldman RA, editors. 8th International Workshop on ‘Campylobacters, Helicobacters and Related Organisms’. New York (NY): Plenum Press; p. 329–334.
- Lammerding AM, Harris JE, Loir H, Muckel CA, Irwin RJ. 1994. An isolation method for Arcobacter butzleri from poultry. Dairy Food Environ Sanitation. 14:600.
- Lappi V, Archer JR, Cebelinski E, Leano F, Besser JM, Klos RF, Medus C, Smith KE, Fitzgerald C, Davis JP. 2013. An outbreak of foodborne illness among attendees of a wedding reception in Wisconsin likely caused by Arcobacter butzleri. Foodborne Pathog Dis. 10:250–255.
- Lau SKP, Woo PCY, Teng J, Leung KW, Yuen KY. 2002. Identification by 16S ribosomal RNA gene sequencing of Arcobacter bacteraemia in a patient with acute gangrenous appendicitis. Mol Pathol. 55:182–185.
- Lee MH, Cheon DS, Choi S, Lee BH, Jung JY, Choi C. 2010. Prevalence of Arcobacter species isolated from retail meats in Korea. J Food Protect. 73:1313–1316.
- Lee MH, Choi, C. 2013. Survival of Arcobacter butzleri in apple and pear purees. J Food Safety. 33:333–339.
- Lehmann D, Alter T, Lehmann L, Uherkova S, Seidler T, Gölz G. 2015. Prevalence, virulence gene distribution and genetic diversity of Arcobacter in food samples in Germany. Berl Munch Tierarztl Wochenschr. 128:163–168.
- Lehner A, Tasara T, Stephan R. 2005. Relevant aspects of Arcobacter spp. as potential food-borne pathogens. Int J Food Microbiol. 102:127–135.
- Lerner J, Brumberger V, Preac-Mursic V. 1994. Severe diarrhea associated with Arcobacter butzleri. Eur J Clin Microbiol Infect Dis. 13:660–662.
- Levican A, Alkeskas A, Günter C, Forsythe SJ, Figueras MJ. 2013b. Adherence to and invasion of human intestinal cells by Arcobacter species and their virulence genotypes. Appl Environ Microbiol. 79:4951–4957.
- Levican A, Collado L, Aguilar C, Yustes C, Dieguez AL, Romalde JL, Figueras MJ. 2012. Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst Appl Microbiol. 35:133–138.
- Levican A, Collado L, Figueras MJ. 2013a. Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst Appl Microbiol. 36:22–27.
- Levican A, Collado L, Figueras MJ. 2016. The use of two culturing methods in parallel reveals a high prevalence and diversity of Arcobacter spp. in a wastewater treatment plant. Biomed Res Int. 2016:8132058.
- Levican A, Collado L, Yustes C, Aguilar C, Figueras MJ. 2014. Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Appl Environ Microbiol. 80:385–391.
- Levican A, Figueras MJ. 2013. Performance of five molecular methods for monitoring Arcobacter spp. BMC Microbiol. 13:220.
- Levican A, Rubio-Arcos S, Martinez-Murcia A, Collado L, José Figueras M. 2015. Arcobacter ebronensis sp. nov. and Arcobacter aquimarinus sp. nov., two new species isolated from marine environment. Syst Appl Microbiol. 38:30–35.
- Logan EF, Neill SD, Mackie DP. 1982. Mastitis in dairy cows associated with an aerotolerant Campylobacter. Vet Res. 110:229–230.
- Manke TR, Wesley IV, Dickson JS, Harmon KM. 1998. Prevalence and genetic variability of Arcobacter species in mechanically separated turkey. J Food Prot. 61:1623–1628.
- Marshall SM, Melito PL, Woodward DL, Johnson WM, Rodgers FG, Mulvey MR. 1999. Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 37:4158–4160.
- Maugeri TL, Irrera GP, Lentini V, Carbone M, Fera MT, Gugliandolo C. 2005. Detection and enumeration of Arcobacter spp. in the coastal environment of the straits of Messina (Italy). New Microbiol. 28:177–182.
- McClung CR, Patriquin DG, Davis RE. 1983. Campylobacter nitrofigilis sp. nov., a nitrogen-fixing bacterium associated with roots of Spartina alterniflora. Int J Syst Bacteriol. 33:605–612.
- McGregor AC, Wright SG. 2015. Gastrointestinal symptoms in travellers. Clin Med. 15:93–95.
- Medina G, Flores-Martin S, Fonseca B, Otth C, Fernandez H. 2014. Mechanisms associated with phagocytosis of Arcobacter butzleri by Acanthamoeba castellanii. Parasitol Res. 113:1933–1942.
- Merga JY, Leatherbarrow AJH, Winstanley C, Bennett M, Hart CA, Miller WG, Williams NJ. 2011. Comparison of Arcobacter isolation methods, and diversity of Arcobacter spp. in Cheshire, United Kingdom. Appl Environ Microbiol. 77:1646–1650.
- Merga JY, Royden A, Pandey AK, Williams NJ. 2014. Arcobacter spp. isolated from untreated domestic effluent. Lett Appl Microbiol. 59:122–126.
- Merga JY, Williams NJ, William G. Miller WG, Leatherbarrow AJH, Bennett M, Hall N, Ashelford KE, Winstanley C. 2013. Exploring the diversity of Arcobacter butzleri from cattle in the UK using MLST and whole genome sequencing. PLoS One. 8:e55240.
- Mesbah M, Premachandran U, Whitman WB. 1989. Precise measurement of the G+C content of deoxyribonucleicacid by high-performance liquid chromatography. Int J Syst Bacteriol. 39:159–167.
- Miller WG, Parker CT, Rubenfield M, Mendz GL, Wösten MM, Ussery DW, Stolz JF, Binnewies TT, Hallin PF, Wang G, et al. 2007. The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One. 2:1358.
- Miller WG, Wesley IV, On SLW, Houf K, Megraud F, Wang G, Yee E, Srijan A, Mason CJ. 2009. First multi-locus sequence typing scheme for Arcobacter spp. BMC Microbiol. 14:196.
- Miller WG, Yee E, Jolley KA, Chapman MH. 2014. Use of improved atpA amplification and sequencing method to identify members of the Campylobacteraceae and Helicobacteraceae. Lett Appl Microbiol. 58:582–590.
- Mohan HV, Rathore RS, Dhama K, Ramees TP, Patyal A, Bagalkot PS, Wani MY, Bhilegaonkar KN, Kumar A. 2014. Prevalence of Arcobacter spp. in humans, animals and foods of animal origin in India based on cultural isolation, antibiogram, PCR and multiplex PCR detection. Asian J Anim Vet Adv. 9:452–466.
- Moreno Y, Alonso JL, Botella S, Ferrus MA, Hernandez J. 2004. Survival and injury of Arcobacter after artificial inoculation into drinking water. Res Microbiol. 155:726–730.
- Moreno Y, Botella S, Alonso JL, Ferrus MA, Hernandez M, Hernandez J. 2003. Specific detection of Arcobacter and Campylobacter strains in water and sewage by PCR and fluorescent in situ hybridization. Appl Environ Microbiol. 69:1181–1186.
- Mottola A, Bonerba E, Bozzo G, Marchetti P, Celano GV, Colao V, Terio V, Tantillo G, Figueras MJ, Di Pinto A. 2016. Occurrence of emerging food-borne pathogenic Arcobacter spp. isolated from pre-cut (ready-to-eat) vegetables. Int J Food Microbiol. 236:33–37.
- Neill SD, Cambell JN, O'brien JJ, Weatherup STC, Ellis WA. 1985. Taxonomic position of Campylobacter cryaerophila sp. Nov. Int J Systemat Bacteriol. 35:342–356.
- Neill SD, Ellis WA, O'brien JJ. 1979. Designation of aerotolerant Campylobacter like organisms from porcine and bovine abortions to the genus Campylobacter. Res Vet Sci. 27:180–186.
- Neubauer J, Jauk V, Szolgyenyi W. 2003. Phenotypic and genotypic differentiation of Campylobacter spp. isolated from Austrian broiler farms: a comparison. Avian Pathol. 32:33–37.
- Neubaure C, Hess M. 2006. Detection and identification of food borne pathogens of the genera Campylobacter, Arcobacter and Helicobacter by multiplex PCR in poultry and poultry products. J Vet Med. 53:376–381.
- Num SM, Useh NM. 2013. Nanotechnology applications in veterinary diagnostics and therapeutics. Sokoto J Vet Sci. 11:10–14.
- Ohlendorf DS, Murano EA. 2002. Prevalence of Arcobacter spp. in raw ground pork from several geographical regions according to various isolation methods. J Food Protect. 65:1700–1705.
- On SLW, Jensen TK, Bille-Hansen V, Jorsal SE, Vandamme P. 2002. Prevalence and diversity of Arcobacter spp. isolated from the internal organs of spontaneous porcine abortions in Denmark. Vet Microbiol. 85:159–167.
- On SLW, Stacey A, Smyth J. 1995. Isolation of Arcobacter butzleri from a neonate with bacteremia. J Infect. 31:225–227.
- Ongor H, Cetinkaya B, Açik MN, Atabay HI. 2004. Investigation of Arcobacters in meat and fecal samples of clinically healthy cattle in Turkey. Lett Appl Microbiol. 38:339–344.
- Ottaviani D, Chierichetti S, Rocchegiani E, Bartolini C, Masini L, Santarelli S, Leoni F. 2013. Bioaccumulation experiments in mussels contaminated with the food-borne pathogen Arcobacter butzleri: preliminary data for risk assessment. Biomed Res Int. 2013:153419.
- Park S, Jung YT, Kim S, Yoon JH. 2016. Arcobacter acticola sp. nov., isolated from seawater on the East Sea in South Korea. J Microbiol. 54:655–659.
- Patyal A, Rathore RS, Mohan HV, Dhama K, Kumar A. 2011. Prevalence of Arcobacter spp. in humans, animals and foods of animal origin including sea food from India. Transbound Emerg Dis. 58:402–410.
- Pejchalova M, Dostali´kova, E. Slamova M, Brozkova I, Vytrasova J. 2008. Prevalence and diversity of Arcobacter spp. in the Czech Republic. J Food Protect. 71:719–727.
- Pentimalli D, Pegels N, Garcia T, Martin R, González I. 2009. Specific PCR detection of Arcobacter butzleri, Arcobacter cryaerophilus, Arcobacter skirrowii and Arcobacter cibarius in chicken meat. J Food Protect. 72:1491–1495.
- Petersen RF, Harrington CS, Kortegaard HE, On SL. 2007 . A PCR-DGGE method for detection and identification of Campylobacter, Helicobacter, Arcobacter and related Epsilobacteria and its application to saliva samples from humans and domestic pets. J Appl Microbiol. 103:2601–2615.
- Phillips CA, Duggan J. 2001. The effect of EDTA and trisodium phosphate, alone and in combination with nisin, on the growth of Arcobacter butzleri in culture. Food Microbiol. 18:547–554.
- Phillips CA, Long C. 2001. The survival of Arcobacter butzleri on chicken. Int J Med Microbiol. 291:93.
- Phillips CA. 1999. The effect of citric acid, lactic acid, sodium citrate and sodium lactate, alone and in combination with nisin, on the growth of Arcobacter butzleri. Lett Appl Microbiol. 29:424–428.
- Phillips CA. 2001. Arcobacter spp in food: isolation, identification, and control. Trends Food Sci Technol. 12:263–275.
- Prouzet-Mauleon V, Labadi L, Bouges N, Menard A, Megraud F. 2006. Arcobacter butzleri: underestimated enteropathogen. Emerg Infect Dis. 12:307–309.
- Quinones B, Parker CT, Janda JM, Miller WG, Mandrell RE. 2007. Detection and genotyping of Arcobacter and Campylobacter isolates from retail chicken samples by use of DNA oligonucleotide arrays. Appl Environ Microbiol. 73:3645–3655.
- Rahimi E. 2014. Prevalence and antimicrobial resistance of Arcobacter species isolated from poultry meat in Iran. Br Poult Sci. 55:174–180.
- Ramees TP, Rathore RS, Bagalakote PS, Mohan HV, Kumar A, Dhama K. 2014a. Multiplex PCR detection of Arcobacter butzleri and Arcobacter cryaerophilus in skin of poultry. J Pure Appl Microbiol. 8:1755–1758.
- Ramees TP, Rathore RS, Bagalkot PS, Mohan HV, Kumar A, Dhama K. 2014b. Detection of Arcobacter butzleri and Arcobacter cryaerophilus in clinical samples of humans and foods of animal origin by cultural and multiplex PCR based methods. Asian J Anim Vet Adv. 9:243–252.
- Ramees TP, Rathore RS, Bagalkot PS, Ravi Kumar GVPPS, Mohan HV, Anoopraj R, Kumar A, Dhama K. 2014c. Real-time PCR detection of Arcobacter butzleri and Arcobacter cryaerophilus in chicken meat samples. J Pure Appl Microbiol. 8:3165–3169.
- Ramees TP, Rathore RS, Bagalkot PS, Sailo Blessa, Mohan HV, Kumar A, Dhama K, Singh RK. 2014d. Genotyping and genetic diversity of Arcobacter butzleri and Arcobacter cryaerophilus isolated from different sources by using ERIC-PCR from India. Vet Quart. 34:211–217.
- Rasmussen LH, Jette K, Jens PC, Hanne I. 2013. Multilocus sequence typing and biocide tolerance of Arcobacter butzleri from Danish broiler carcasses. BMC Res Notes. 6:322.
- Revez J, Huuskonen M, Ruusunen M, Lindström M, Hänninen ML. 2013. Arcobacter species and their pulsed-field gel electrophoresis genotypes in Finnish raw milk during summer 2011. J Food Prot. 76:1630–1632.
- Ricci JCD, Hernandez ME. 2000. Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol. 20:79–108.
- Rice EW, Rodgers MR, Wesley IV, Johnson CH, Tanner SA. 1999. Isolation of Arcobacter butzleri from ground water. Lett Appl Microbiol. 28:31–35.
- Rivas L, Fegan N, Vanderlinde P. 2004. Isolation and characterisation of Arcobacter butzleri from meat. Int J Food Microbiol. 91:31–41.
- Romero J, Varela GM, Laclette JP, Espejo RT. 2002. Bacterial 16S rRNA gene analysis revealed that bacteria related to Arcobacter spp. constitute an abundant and common component of the oyster microbiota (Tiostrea chilensis). Microb Ecol. 44:365–371.
- Ruiz N. 2008. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA. 105:15553–15557.
- Salas-Massó N, Andree KB, Furones MD, Figueras MJ. 2016. Enhanced recovery of Arcobacter spp. using NaCl in culture media and re-assessment of the traits of Arcobacter marinus and Arcobacter halophilus isolated from marine water and shellfish. Sci Total Environ. 566–567:1355–1361.
- Samarajeewa AD, Hammad A, Masson L, Khan IU, Scroggins R, Beaudette LA. 2015. Comparative assessment of next-generation sequencing, denaturing gradient gel electrophoresis, clonal restriction fragment length polymorphism and cloning-sequencing as methods for characterizing commercial microbial consortia. J Microbiol Methods. 108:103–111.
- Samie A, Obi CL, Barrett LJ, Powell SM, Guerrant RL. 2007. Prevalence of Campylobacter species, Helicobacter pylori and Arcobacter species in stool samples from the Venda region, Limpopo, South Africa: studies using molecular diagnostic methods. J Infect. 54:558–566.
- Sasi Jyothsna TS, Rahul K, Ramaprasad EV, Sasikala Ch, Ramana ChV. 2013. Arcobacter anaerophilus sp. nov., isolated from an estuarine sediment and emended description of the genus Arcobacter. Int J Syst Evol Microbiol. 63:4619–4625.
- Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States – unspecified agents. Emerg Infect Dis. 17:16.
- Scarano C, Giacometti F, Manfreda G, Lucchi A, Pes E, Spanu C, De Santis EP, Serraino A. 2014. Arcobacter butzleri in sheep ricotta cheese at retail and related sources of contamination in an industrial dairy plant. Appl Environ Microbiol. 80:7036–7041.
- Schmitt B, Henderson L. 2005. Diagnostic tools for animal diseases. Rev Sci Tech. 24:243–250.
- Scullion R, Harrington CS, Madden RH. 2004. A comparison of three methods for the isolation of Arcobacter spp. from retail raw poultry in Northern Ireland. J Food Prot. 67:799–804.
- Serraino A, Florio D, Giacometti F, Piva S, Mion D, Zanoni RG. 2013b. Presence of Campylobacter and Arcobacter species in in-line milk filters of farms authorized to produce and sell raw milk and of a water buffalo dairy farm in Italy. J Dairy Sci. 96:2801–2807.
- Serraino A, Giacometti F, Daminelli P, Losio MN, Finazzi G, Marchetti G, Zambrini AV, Rosmini R. 2013a. Survival of Arcobacter butzleri during production and storage of artisan water buffalo mozzarella cheese. Foodborne Pathog Dis. 10:820–824.
- Serraino A, Giacometti F. 2014. Short communication: occurrence of Arcobacter species in industrial dairy plants. J Dairy Sci. 97:2061–2065.
- Shah AH, Saleha AA, Murugaiyah M, Zunita Z, Memon AA. 2012a. Prevalence and distribution of Arcobacter spp. in raw milk and retail raw beef. J Food Prot. 75:1474–1478.
- Shah AH, Saleha AA, Zunita Z, Cheah YK, Murugaiyah M, Korejo NA. 2012b. Genetic characterization of Arcobacter isolates from various sources. Vet Microbiol. 160:355–361.
- Shah AH, Saleha AA, Zunita Z, Murugaiyah M, Aliyu AB, Jafri N. 2013. Prevalence, distribution and antibiotic resistance of emergent Arcobacter spp. from clinically healthy cattle and goats. Transbound Emerg Dis. 60:9–16.
- Shah AH, Saleha AA, Zunita Z, Murugaiyah M. 2011. Arcobacter– an emerging threat to animals and animal origin food products? Tr Food Sci Technol. 22:225–236.
- Shirzad Aski H, Tabatabaei M, Khoshbakht R, Raeisi M. 2016. Occurrence and antimicrobial resistance of emergent Arcobacter spp. isolated from cattle and sheep in Iran. Comp Immunol Microbiol Infect Dis. 44:37–40.
- Silipo A, Sturiale L, Perino V, Garozzo D, Lanzetta R, Parrilli M, Molinaro A. 2010. The structure of the carbohydrate backbone of the lipooligosaccharide from the halophilic bacterium Arcobacter halophilus. Carbohydr Res. 345:850–853.
- Skrivanová E, Molatová Zuzana, Matenová Michaela, Houf Kurt, Marounek Milan. 2011. Inhibitory effect of organic acids on arcobacters in culture and their use for control of Arcobacter butzleri on chicken skin. Int J Food Microbiol. 144:367–371.
- Snelling WJ, Matsuda M, Moore JE, Dooley JS. 2006. Under the microscope: Arcobacter. Lett Appl Microbiol. 42:7–14.
- Son I, Englen MD, Berrang ME, Fedorka-Carry PJ, Harrison MA. 2007. Prevalence of Arcobacter and Campylobacter on broiler carcases during processingInt. J Food Microbiol. 113:16–22.
- Son I, Englen MD, Berrang ME, Fedorka-Cray PJ, Harrison MA. 2006. Genetic diversity of Arcobacter and Campylobacter on broiler carcasses during processing. J Food Prot. 69:1028–1033.
- Stirling J, Griffith M, Blair I, Cormican M, Dooley JS, Goldsmith CE, Glover SG, Loughrey A, Lowery CJ, Matsuda M, et al. 2008. Prevalence of gastrointestinal bacterial pathogens in a population of zoo animals. Zoonoses Public Health. 55:166–172.
- Strugala V, Allen A, Dettmar PW, Pearson JP. 2003. Colonic mucin: methods of measuring mucus thickness. P Nutr Soc. 62:237–243.
- Suarez DL, Wesley IV, Larson DJ. 1997. Detection of Arcobacter species in gastric samples from swine. Vet Microbiol. 57:325–336.
- Sudhakar NR, Manjunathachar HV, Karthik K, Sahu S, Gopi M, Shanthaveer SB, Nagaraja KH, Shinde S, Tamilmahan P, Madhu DN, Maurya PS. 2013. RNA interference in parasites: prospects and pitfalls. Adv Anim Vet Sci. 1:1–6.
- Suelam II A. 2012. Isolation and identification of Arcobacter species recovered from rabbits in Zagazig, Egypt. Int J of Micr Res. 3:87–92.
- Tabatabaei M, Aski HS, Shayegh H, Khoshbakht R. 2014. Occurrence of six virulence-associated genes in Arcobacter species isolated from various sources in Shiraz, Southern Iran. Microb Pathog. 66:1–4.
- Talay F, Molva C, Atabay HI. 2016. Isolation and identification of Arcobacter species from environmental and drinking water samples. Folia Microbiol. 61:479–484. doi:10.1007/s12223-016-0460-0
- Tee W, Baird R, Dyall-Smith M, Dwyer B. 1988. Campylobacter cryaerophila isolated from a human. J Clin Microbiol. 26:2469–2473.
- Tiwari R, Chakraborty S, Dhama K, Rajagunalan S, Singh SV. 2013. Antibiotic resistance – an emerging health problem: causes, worries, challenges and solutions – a review. Int J Curr Res. 5:1880–1892.
- Tiwari R, Dhama K, Chakraborty S, Kumar A, Rahal A, Kapoor S. 2014. Bacteriophage therapy for safeguarding animal and human health: a review. Pak J Biol Sci. 17:301–315.
- Tiwari R, Karthik K, Rana R, Malik YS, Dhama K, Joshi SK. 2016. Quorum sensing inhibitors/antagonists countering food spoilage bacteria-need molecular and pharmaceutical intervention for protecting current issues of food safety. Int J Pharmacol. 12:262–271.
- Tsang RS, Luk JM, Woodward DL, Johnson WM. 1996. Immunochemical characterization of a haemagglutinating antigen of Arcobacter spp. FEMS Microbiol Lett. 136:209–213.
- Ünver A, Atabay Hİ, Şahin M, Çelebi Ö. 2013. Antimicrobial susceptibilities of various Arcobacter species. Turk J Med Sci. 43:548–552.
- Van Driessche E, Houf K, Van Hoof J, De Zutter L, Vandamme P. 2003. Isolation of Arcobacter species from animal feces. FEMS Microbiol. 229:243–248.
- Van Driessche E, Houf K, Vangroenweghe F, De Zutter L, Van Hoof J. 2005. Prevalence, enumeration and strain variation of Arcobacter species in the faeces of healthy cattle in Belgium. Vet Microbiol. 105:149–154.
- Van Driessche E, Houf K, Vangroenweghe F, Nollet N, De Zutter L, Vandamme P, Van Hoof J. 2004. Occurrence and strain diversity of Arcobacter species isolated from healthy Belgian pigs. Res Microbiol. 155:662–666.
- Van Driessche E, Houf K. 2007a. Characterization of the Arcobacter contamination on Belgian pork carcasses and raw retail pork. Int J Food Microbiol. 118:20–26.
- Van Driessche E, Houf K. 2007b. Discrepancy between the occurrence of Arcobacter in chickens and broiler carcass contamination. Poult Sci. 86:744–751.
- Vandamme P, De Ley J. 1991. Proposal for a new family Campylobacteraceae. Int J Syst Bacteriol. 41:451–455.
- Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. 1991. Revision of Campylobacter, H elicobacter and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter genera. Int J Syst Bacteriol. 41:88–103.
- Vandamme P, Giesendorf AJ, Van Belkum P, Lauwers S, Kersters K, Butzler P, Goossens H, Quint WG. 1993. Discrimination of epidemic and sporadic isolates of Arcobacter butzleri by polymerase chain reaction-mediated DNA fingerprinting. J Clin Microbiol. 31:3317–3319.
- Vandamme P, Pugina P, Benzi G, Van Etterijck R, Vlaes L, Kersters K, Butzler JP, Lior H, Lauwers S. 1992a. Outbreak of recurrent abdominal cramps associated with Arcobacter butzleri in an Italian school. J Clin Microbiol. 30:2335–2337.
- Vandamme P, Vanvanneyt M, Pot B, Mels L, Hoste B, Dewettinck D, Vlaes L, Van Den Borre C, Higgins R, Hommez J, et al. 1992b. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. Nov. and Arcobacter skirrowii sp. Nov., and aerotolerant bacterium isolated from veterinary specimens. Int J Syst Bacteriol. 42:344–356.
- Van den Abeele AM, Vogelaers D, Van Hende J, Houf K. 2014. Prevalence of Arcobacter species among humans, Belgium, 2008–2013. Emerg Infect Dis. 20:1731–1734.
- Van den Abeele AM, Vogelaers D, Vanlaere E, Houf K. 2016. Antimicrobial susceptibility testing of Arcobacter butzleri and Arcobacter cryaerophilus strains isolated from Belgian patients. J Antimicrob Chemother. 71:1241–1244.
- Vandenberg O, Dediste A, Houf K, Ibekwem S, Souayah H, Cadranel S, Douat N, Zissis G, Butzler JP, Vandamme P. 2004. Arcobacter species in humans. Emerg Infect Dis. 10:1863–1867.
- Vergis J, Negi M, Poharkar K, Das DP, Malik SV, Kumar A, Doijad SP, Barbuddhe SB, Rawool DB. 2013. 16S rRNA PCR followed by restriction endonuclease digestion: a rapid approach for genus level identification of important enteric bacterial pathogens. J Microbiol Methods. 95:353–356.
- Vilar MJ, Pena FJ, Pe´rez I, Die´guez FJ, Sanjuan ML, Rodri´guez-Otero JL, Yus E. 2010. Presence of Listeria, Arcobacter, and Campylobacter spp. in dairy farms in Spain. Berl Munch Tierarztl Wochenschr. 23:58–62.
- Villalobos EG, Jaramillo HF, Ulate CC, Echandi ML. 2013. Isolation and identification of zoonotic species of genus arcobacter from chicken viscera obtained from retail distributors of the metropolitan area of San José, Costa Rica. J Food Prot. 76:879–882.
- Villanueva MP, Medina G, Fernández H. 2016. Arcobacter butzleri survives within trophozoite of Acanthamoeba castellanii. Rev Argent Microbiol. 48:105–109.
- Villarruel-López A, Márquez-González M, Garay-Martínez LE, Zepeda H, Castillo A, Mota de la Garza L, Murano EA, Torres-Vitela R. 2003. Isolation of Arcobacter spp. from retail meats and cytotoxic effects of isolates against vero cells. J Food Prot. 66:1374–1378.
- Vytrasová J, Pejchalová M, Harsová K, Bínová S. 2003. Isolation of Arcobacter butzleri and A. cryaerophilus in samples of meats and from meat-processing plants by a culture technique and detection by PCR. Folia Microbiol. 48:227–232.
- Wang X, Seo DJ, Lee MH, Choi C. 2014. Comparison of conventional PCR, multiplex PCR and loop-mediated isothermal amplification assays for rapid detection of Arcobacter species. J Clin Microbiol. 52:557–563.
- Webb AL, Boras VF, Kruczkiewicz P, Selinger LB, Taboada EN, Inglis GD. 2016. Comparative detection and quantification of Arcobacter butzleri in stools from diarrheic and nondiarrheic people in Southwestern Alberta, Canada. J Clin Microbiol. 54:1082–1088.
- Webb AL, Kruczkiewicz P, Selinger LB, Inglis GD, Taboada EN. 2015. Development of a comparative genomic fingerprinting assay for rapid and high resolution genotyping of Arcobacter butzleri. BMC Microbiol. 15:94.
- Wesley IV, Baetz AL. 1999. Natural and experimental infections of Arcobacter in poultry. Poult Sci. 78:536–545.
- Wesley IV, Schroder-Tucker L, Baetz AL, Dewhurst FE, Paster BJ. 1995. Arcobacter butzleri-specific 16S rRNA based probes. J Clin Microbiol. 33:1691–1698.
- Wesley IV, Schroeder-Tucker L, Franklin SL. 2003. Recovery of Arcobacter species from exotic animal species. Int J Med Microbiol. 293:57.
- Wesley IV, Wells SJ, Harmon KM, Green A, Schroeder-Tucker L, Glover M, Siddique I. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl Environ Microbiol. 66:1994–2000.
- Wesley IV. 1994. Arcobacter infections. In: Beran GW, Steele JH, editors. In handbook of zoonoses. CRC Press; p. 181–190.
- Wesley IV. 1997. Helicobacter and Arcobacter: potential human foodborne pathogens? Trends Food Sci Technol. 8:293–299.
- Whiteduck-Léveillée J, Cloutier M, Topp E, Lapen DR, Talbot G, Villemur R, Khan IU. 2016a. Development and evaluation of multiplex PCR assays for rapid detection of virulence-associated genes in Arcobacter species. J Microbiol Methods. 121:59–65.
- Whiteduck-Léveillée K, Whiteduck-Léveillée J, Cloutier M, Tambong JT, Xu R, Topp E, Arts MT, Chao J, Adam Z, Lévesque CA, et al. 2015. Arcobacter lanthieri sp. nov., isolated from pig and dairy cattle manure. Int J Syst Evol Microbiol. 65:2709–2716.
- Whiteduck-Léveillée K, Whiteduck-Léveillée J, Cloutier M, Tambong JT, Xu R, Topp E, Arts MT, Chao J, Adam Z, Lévesque CA, et al. 2016b. Identification, characterization and description of Arcobacter faecis sp. nov., isolated from a human waste septic tank. Syst Appl Microbiol. 39:93–99.
- Winters DK, Slavik MF. 2000. Multiplex PCR detection of Campylobacter jejuni and Arcobacter butzleri in food products. Mol Cell Probes. 14:95–99.
- Wirsen CO, Sievert SM, Cavanaugh CM, Molyneaux SJ, Ahmad A, Taylor LT, DeLong EF, Taylor CD. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl Environ Microbiol. 68:316–325.
- Woo PCY, Chong KTK, Leung KW, Que TL, Yuen KY. 2001. Identification of Arcobacter cryaerophilus isolated from a traffic accident victim with bacteremia by 16S ribosomal RNA gene sequencing. Diagn Microbiol Infect Dis. 40:125–127.
- Wybo I, Breynaert J, Lauwers S, Lindenburg F, Houf K. 2004. Isolation of Arcobacter skirrowii from a patient with chronic diar-rhea. J Clin Microbiol. 42:1851–1852.
- Yadav AS, Kolluri G, Gopi M, Karthik K, Malik YS, Dhama K. 2016. Exploring alternatives to antibiotics as health promoting agents in poultry- a review. J Exp Biol Agri Sci. 4:368–383.
- Yan JJ, Ko WC, Huang AH, Chen HM, Jin YT, Wu JJ. 2000. Arcobacter butzleri bacteremia in a patient with liver cirrhosis. J Formos Med Assoc. 99:166–169.
- Yesilmen S, Vural A, Erkan ME, Yildirim IH. 2014. Prevalence and antimicrobial susceptibility of Arcobacter species in cow milk, water buffalo milk and fresh village cheese. Int J Food Microbiol. 188:11–14.
- Yildiz H, Aydin S. 2006. Pathological effects of Arcobacter cryaerophilus infection in rainbow trout (Oncorhynchus mykissWalbaum). Acta Vet Hung. 54:191–199.
- Zacharow I, Bystroń J, Wałecka-Zacharska E, Podkowik M, Bania J. 2015. Prevalence and antimicrobial resistance of Arcobacter butzleri and Arcobacter cryaerophilus isolates from retail meat in Lower Silesia region, Poland. Pol J Vet Sci. 18:63–69.
- Zhang Z, Yu C, Wang X, Yu S, Zhang XH. 2016. Arcobacter pacificus sp. nov., isolated from seawater of the South Pacific Gyre. Int J Syst Evol Microbiol. 66:542–547.
- Zur Bruegge J, Backes C, Gölz G, Hemmrich-Stanisak G, Scharek-Tedin L, Franke A, Alter T, Einspanier R, Keller A, Sharbati S. 2016. MicroRNA response of primary human macrophages to Arcobacter butzleri infection. Eur J Microbiol Immunol. 6:99–108.
- Zur Bruegge J, Hanisch C, Einspanier R, Alter T, Gölz G, Sharbati S. 2014. Arcobacter butzleri induces a pro-inflammatory response in THP-1 derived macrophages and has limited ability for intracellular survival. Int J Med Microbiol. 304:1209–1217.