Abstract
Diatom species identification with DNA metabarcoding is an economical, fast and reliable alternative to identification via light microscopy for river quality monitoring. Using a short DNA sequence of the rbcL gene and ‘Diat.barcode’, a reference barcode library, enables the identification of more than 90% of the environmental sequences to species level in French rivers. But the completeness of this library is much lower in other regions, such as the tropical French overseas departments. A barcode library completion method using high-throughput sequencing data combined with microscopy count data from natural samples (Rimet et al. Citation2018) was applied and tested in rivers of Martinique and Guadeloupe (West Indies), for which only 45% of the environmental sequences could be identified to species level using Diat.barcode v9. Assigning barcodes to the most abundant species in the islands by this method is illustrated with Ulnaria goulardii and two new species belonging to Nupela and Epithemia, which are also described in this paper. The more complex situation of morphologically similar species is illustrated by reference to Gomphonema designatum and G. bourbonense. Using a combination of molecular and morphological data, their conspecificity, as G. bourbonense, is demonstrated with their reference barcodes. However, when several morphologically similar species and several environmental sequences belonging to the same clade are present, it is not possible to relate the barcodes to corresponding morphological species.
Applying this method enabled the Diat.barcode library (v.10) to be updated, with 84% of the environmental sequences from the West Indies now identifiable at the species level. However, many morphological species still lack barcodes. In these cases, more classical methods, such as cell isolation, Sanger sequencing and morphological observations of cultures, must be applied.
Introduction
Given their value as ecological indicators, diatoms are now required for the assessment of water ecosystems quality in Europe (e.g. Kelly et al. Citation2014) as part of the Water Framework Directive (European Commission, Citation2000) and in the USA under the Clean Water Act (e.g. Barbour et al. Citation1999, Potapova & Charles Citation2007, Hausmann et al. Citation2016). To apply diatoms as monitoring tools, species have to be identified and counted and then biotic indices calculated (Rimet Citation2012). These methods are standardized, and identification and counting are carried out using microscopy (CEN Citation2014). However, there are some difficulties to identify species with light microscopy. It requires time and trained analysts with good knowledge of the taxonomic literature, but inter-analyst variation can often influence the final result (Kahlert et al. Citation2009). In addition, many semi-cryptic species and species complexes (e.g. Trobajo et al. Citation2009, Kermarrec et al. Citation2012, Abarca et al. Citation2014, Kelly et al. Citation2015, Pinseel et al.Citation2020) make morphological identification complex.
An alternative solution is the DNA barcoding approach (Hebert et al. Citation2005), in which specific short DNA sequences, so-called barcodes, enable diatom species to be identified. DNA metabarcoding (Pompanon et al. Citation2011), based on High-Throughput-Sequencing (HTS), expanded the barcoding concept to environmental samples, in which species mixtures can be identified from natural samples. Several studies have successfully applied this approach to diatoms (Kermarrec et al. Citation2013, Visco et al. Citation2015, Zimmermann et al. Citation2015, Vasselon et al. Citation2017a, Rivera et al. Citation2018a, Citationb). To enable efficient, accurate species identification based on DNA barcodes, reliable reference barcoding libraries are required. These libraries connect DNA barcodes to species and are used to identify the species present in samples. Several curated libraries for protists exist (e.g. PR2 for microbial diversity in Guillou et al. [Citation2013], Phytool for phytoplankton in Canino et al. [Citation2021]) and for diatoms Diat.barcode, which is open access and has been maintained since 2012 (Rimet et al. Citation2019). Reference barcodes in Diat.barcode come from two sources: (1) the NCBI nucleotide database and (2) unpublished sequences of culture collections. The chosen barcode for this reference library is rbcL, a chloroplast gene marker suitable for species-level identification of diatoms (Kermarrec et al. Citation2013, Citation2014). The last version of the Diat.barcode reference library (v9) contained 8066 sequences from 1491 species in 300 genera. Diat.barcode is almost complete for some regions of Europe, like France, where 91% of the environmental sequences from rivers can be identified to species level (Rimet et al. Citation2021). However, this library is still largely incomplete for the French tropical region, where diatom biomonitoring also must be applied for routine river assessment. This was the case for the French West Indies (Guadeloupe and Martinique islands), where only 45% of the environmental sequences from samples from the river monitoring network in 2018 and 2019 could be identified with Diat.barcode (Rimet et al. Citation2019). There is, therefore, a need to expand this reference database if metabarcoding will be used for diatom monitoring in rivers.
There are several methods for filling diatom reference libraries (Rimet et al. Citation2018): (1) single-cell isolation and culturing, followed by Sanger sequencing (e.g. Evans et al. Citation2007, Trobajo et al. Citation2009, Abarca et al. Citation2014, Zimmermann et al. Citation2014), (2) single-cell PCR followed by microscope observation of the living or oxidized frustule for species identification (Takano & Horiguchi Citation2006, Gomez et al. Citation2012, Hamilton et al. Citation2015, Khan–Bureau et al. Citation2016, Lefebvre et al Citation2017, Skibbe et al. Citation2018, Hamilton et al. Citation2019), (3) direct Sanger sequencing of environmental samples with very low diversity, such as a Didymosphenia M. Schmidt bloom in mountain streams (Jaramillo et al. Citation2015), (4) use of HTS data from environmental samples and comparison to light microscopy (LM) and scanning electron microscopy (SEM) analyses to identify barcodes for target species (Rimet et al. Citation2018). This last method has the advantage that HTS enables sequencing of several hundreds of samples in a single run, generating several millions of sequences at reasonable cost and with good sequencing quality (Pfeiffer et al. Citation2018). However, linking the sequences to the diatom species observed in LM and SEM remains the main challenge.
This last approach was used in this study, with several examples from Guadeloupe and Martinique, where river samples were sequenced with Illumina MiSeq and observed with LM and SEM. The objective of the study is to show that this approach is easily applicable in some cases, even if species are new to science. However, there are more complex situations, and even some for which this approach is not applicable. We analyse these various cases and highlight the reasons for the success or failure of the approach.
Materials and methods
Study site: Martinique (1128 km2) is located in the volcanic arc of the Lesser Antilles, in the Caribbean Sea, between Dominica to the north and Saint Lucia to the south. Guadeloupe (1436 km2) is located in the Caribbean archipelago, between the Tropic of Cancer and the equator. These islands have a tropical climate with two distinct seasons, a generally dry season from December to June and a wet winter season from July to December.
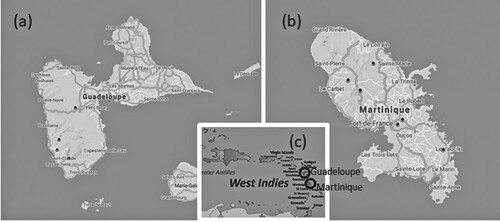
Sampling: The sampling was carried out in 2018 and 2019 during the dry season. One hundred and one samples were collected from the two study areas. The samples were collected from March to June (for Martinique: 12–16 May 2018 and 25–28 March 2019 and 24 June 2019; for Guadeloupe: 15–23 June 2018 and 10–17 April 2019). The sampling procedure followed European standards (European Committee for Standardization Citation2014a, Citation2014b). Benthic diatoms were collected from at least five stones in fast-flowing parts of rivers. The upper surfaces of the stones were scrubbed with a clean toothbrush to collect biofilms. The samples were then fixed with ethanol (final concentration > 70%) following the European protocol for subsequent microscopic and metabarcoding analyses (CEN Citation2014, Citation2018). Fig. 1 shows the sampling site locations.
Fig. 1. Maps of Guadeloupe (a), Martinique (b) and general location in the West Indies (c). The location of the sampling sites is marked with black dots, and the locations of the islands are marked with open circles.
DNA extraction and PCR amplification: DNA extraction was done from the pellet obtained following centrifugation of the biofilm (30 min to 17,000 g) using the NucleoSpin Soil kit according to the manufacturer's instructions, as described by Vasselon et al. (Citation2017b). A small DNA fragment (312 base pairs in length) of rbcL was used for DNA amplification. PCR amplification of the DNA barcode was performed for each sample using a mix of three forward and two reverse primers. The forward primer combined an equimolar mix of Diat_rbcL_708F_1 (AGGTGAAGTAAAAGGTTCWTACTTAAA), Diat_rbcL_708F_2 (AGGTGAAGTTAAAGGTTCWTAYTTAAA) and Diat_rbcL_708F_3 (AGGTGAAACTAAAGGTTCWTACTTAAA); the reverse primer combined an equimolar mix of R3_1 (CCTTCTAATTTACCWACWACTG) and R3_2 (CCTTCTAATTTACCWACAACAG) (Vasselon et al. Citation2017b). Each DNA extract was amplified in triplicate using equimolar mixes of the three forward and two reverse primers. Half the P5 (CTTTCCCTACACGACGCTCTTCCGATCT) and P7 (GGAGTTCAGACGTGTGCTCTTCCGATCT) Illumina adapters were included to the 5′ part of the rbcL forward and reverse primers, respectively. Additionally, blank samples using water were run in parallel to check for potential contamination. Amplifications were performed in a final volume of 25 μL following mix and reaction conditions used previously, the number of amplifications was set to 33 and the conditions of a cycle were as follow: 95°C – 1 min, 54°C – 1 min, 72°C – 1 min (Keck et al. Citation2018).
High-throughput sequencing and bioinformatics processing: The PCR amplicons were purified and used as templates in a second PCR that used Illumina tailed primers targeting the P5 (CTTTCCCTACACGACGCTCTTCCGATCT) and P7 (GGAGTTCAGACGTGTGCTCTTCCGATCT) Illumina adapters. Finally, all generated PCR amplicons were indexed and pooled into a single tube. The final pool was sequenced in GeT-Plage (Toulouse, France) Illumina MiSeq platform using the V2 paired-end sequencing kit (250 bp × 2).
Demultiplexing and adaptors’ removal were performed by the sequencing platform. Further bioinformatic treatment was performed with R software (3.6.1, R development core team). The software package DADA2 version 1.18.0 (Callahan et al. Citation2016) was used with parameters adapted to diatom metabarcoding data available on Github (https://github.com/fkeck/DADA2_diatoms_pipeline). The following bioinformatic steps were carried out: (i) primers were removed using cutadapt (version 2.1); (ii) to keep only good quality sequences (we checked fastqc to have Phred scores above 35), the R1 and R2 reads were truncated to 200 and 170 nucleotides respectively; (iii) only R1 and R2 reads with zero ambiguities and a maximum of two expected errors were kept; (iv) after dereplication, high-quality amplicon sequence variants (ASVs) were selected based on the error rates model and paired reads were merged into one sequence (Tapolczai et al. Citation2019). The last step (iv) cleaned the data by eliminating chimeric sequences. A total of 1.9 million reads were obtained from a single run of good quality, which comparable to the number of reads obtained in earlier studies (e.g. Rivera et al. Citation2020), ranging from 2916 to 33192 (average 19533 reads).
To assign taxa to each DNA sequence, the sequences were compared with the reference library Diat.barcode, version 9 (Rimet et al. Citation2019) using a Naïve Bayesian method with a confidence level of 60% (Youn and Wang Citation2008). All non-diatom sequences (i.e. not assigned to the ‘Bacillariophyta’ phylum) were removed. The data were rarified using rrarefy function (R package vegan, Oksanen et al. Citation2013) according to the number of reads per sample. The length of the sequences kept after these different selection steps was 263 bp (without primers).
Microscopy: The procedure for slide preparation followed French and European standards (Afnor Citation2003, Citation2016). Diatom valves were cleaned using 40% H2O2 and 40% HCl. Cleaned valves were mounted in resin (Naphrax©). Diatoms were counted as described in the NF T 90-354 standard (Afnor Citation2016). Specialized floras were used for identification to the lowest taxonomic level, such as diatom floras of the French West Indies (Eulin et al. Citation2017a, Citationb, Citationc, Citationd, Citatione, Citationf).
Phylogenetic analyses: We constructed a constrained phylogeny by placing the environmental ASVs in a backbone phylogeny. To this end, we first used the alignment available in Diat.barcode v9 (Rimet et al. Citation2019). From this alignment, we selected all long sequences (as noted in Diat.barcode). A total of 3265 sequences were included in the alignment (alignment is available in Diat.barcode file at doi:10.15454/TOMBYZ). The best substitution model was then tested in MEGA7 (Kumar et al. Citation2016). A maximum likelihood (ML) tree was calculated following the best substitution model (GTR + G + I model) with raxmlGUI 2.0 (Stamatakis et al. Citation2005, Silvestro & Michalak Citation2012). We then added the 100 most abundant ASV sequences a posteriori in the phylogeny. This was also done in raxmlGUI under the ‘enforce constraint menu’ and ‘use multifurcating constraint’. One hundred bootstraps were run, ML and a bootstrap search took 27.26 h with an Intel® Core™ i5-8250U CPU @ 1.60 GHz. The trees presented in the results are parts extracted from this tree and represented using MEGA7.
BLAST analyses: Nucleotide BLAST (Basic Local Alignment Search Tool, blastn) was used to identify homologous sequences (Altschul et al. Citation1990). To this end, we used the local Blast tool proposed in BioEdit version 7.2.5 (Hall Citation1999). The nucleotide database we used was Diat.barcode v9 (all rbcL sequences), and the maximum e-values used to determine the sequences were e-110.
Correlations between number of ASV and frustule counts: For each unidentified ASV, correlations between abundances of the ASV sequences (reads number) and the abundances of the suspected species counts from the corresponding study sites were carried out using the Pearson correlation coefficient.
Results
According to the degree of difficulty with which we could establish the correspondence between sequence and morphology, three categories were distinguished: (1) simple cases (ASV2-Ulnaria goulardii, ASV22-Nupela sp. nov., ASV38-Epithemia sp. nov.), (2) intermediate cases (ASV11-Gomphonema bourbonense), (3) complicated cases (ASV5, ASV8, ASV37-Nitschia group). These sequences and their corresponding material (raw material, treated material, slides), as well as their metadata, are registered in the TCC collection and in Genbank (NCBI). All data are also available in Diat.barcode v10.
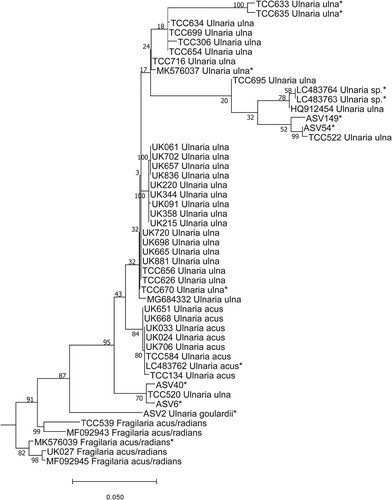
Simple cases: The first undetermined sequence (ASV2) was identified to family level, namely Fragilariaceae, with a bootstrap value of 97% using DADA2 . BLAST identified the sequence as Fragilaria gracilis Østrup, with 95% identity. This guided us to the genus we should look for in the LM slides. The position of ASV2 in the phylogenetic tree was inside the Ulnaria (Kützing) Compère clade (Fig. 2). Checking the slides in LM (Figs 3–11) showed that ASV2 corresponded to Ulnaria goulardii (Brébisson ex Cleve & Grunow) D. M. Williams, Potapova & C. E. Wetzel, a species recently transferred from Fragilaria Lyngbye to Ulnaria (Wetzel et al. Citation2022), and the only Ulnaria species observed by LM in the samples. ASV2 read numbers and frustule counts of U. goulardii showed a significant correlation (R2 = 0.42, p < 0.05). Our results also confirm the transfer to Ulnaria proposed by Wetzel et al. (Citation2022).
Fig. 2. Phylogenetic position of Ulnaria goulardii (ASV2) in the ML tree. Bootstrap values are given for each node and the scale bar gives the number of substitutions per site. ‘*’ indicates sequences added in the phylogeny using the multifurcating constraint.
Figs 3–11. LM micrographs of Ulnaria goulardii. Figs 3–11. Acc. No. TCC1083. From site called Grande Rivière – Vieux habitants amount in Guadeloupe. Scale bar = 10 µm.
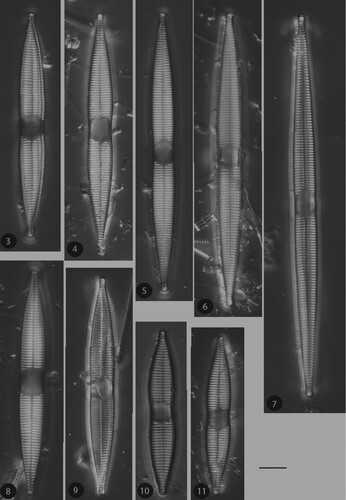
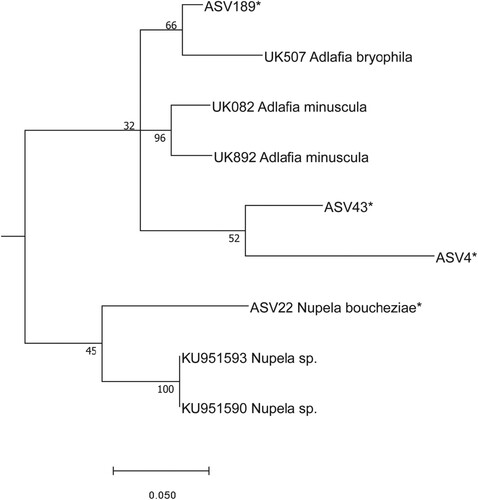
Fig. 12. Phylogenetic position Nupela boucheziae sp. nov. (ASV22) in the ML tree. Bootstrap values are given for each node and the scale bar gives the number of substitutions per site. ‘*’ indicates sequences added in the phylogeny using the multifurcating constraint.
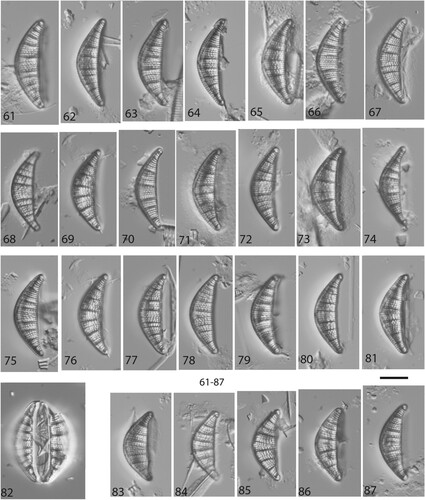
Figs 13–53. LM micrographs of Nupela boucheziae sp. nov. Figs 13–33. Type material Acc. No. TCC1086. Figs 13–22. Valves (with raphe) and Figs. 23–33. Valves (without raphe), from site called Blanche-Pont de l'Alma in Martinique. Figs 34–42. Valves (with raphe) and Figs. 43–53 Valves (rapheless), from river Rivière Bras David in Guadeloupe, Acc. No. TCC1088 (paratype). Scale bar = 10 µm.
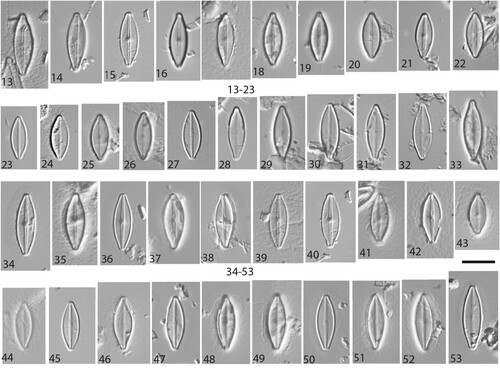
Figs 54–59. SEM micrographs of Nupela boucheziae sp. nov. from type material TCC1086. Figs 54–57. External valve view. Figs. 58–59 Internal valve view. Scale bars = 5 µm (54, 57–59). Scale bars = 2 µm (55, 56).
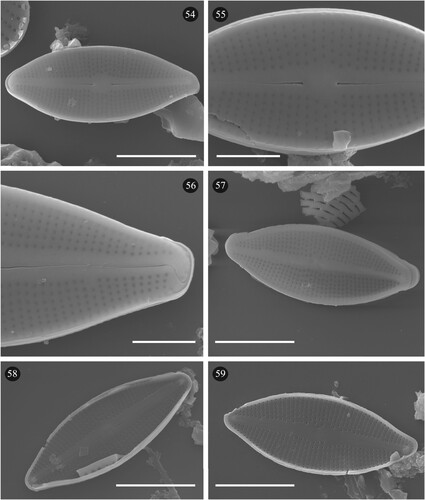
Fig. 60. Phylogenetic position Epithemia boucheziae sp. nov. (ASV38) in the ML tree. Bootstrap values are given for each node and the scale bar gives the number of substitutions per site. ‘*’ indicates sequences added in the phylogeny using the multifurcating constraint.
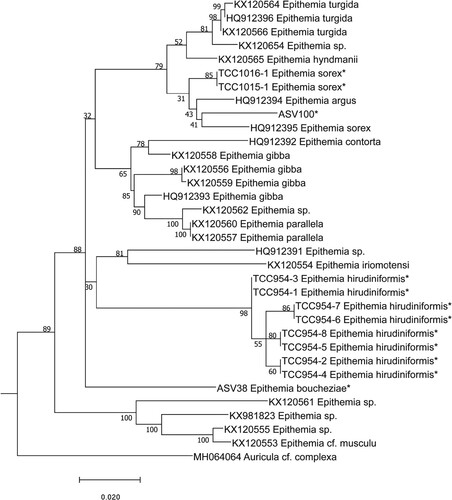
Figs 61–87. LM micrographs of Epithemia boucheziae sp. nov. Figs 61–87. Type material Acc. No. TCC1089. Figs 61–87. from river Rivière Bras David in Guadeloupe. Scale bar = 10 µm.
Figs 88–93. SEM micrographs of Epithemia boucheziae sp. nov. from type material TCC1089. Figs 88–91. External valve view. Figs 92–93. Internal valve view. Scale bars = 10 µm (88, 89, 92). Scale bars = 5 µm (90, 93). Scale bars = 2 µm (91).
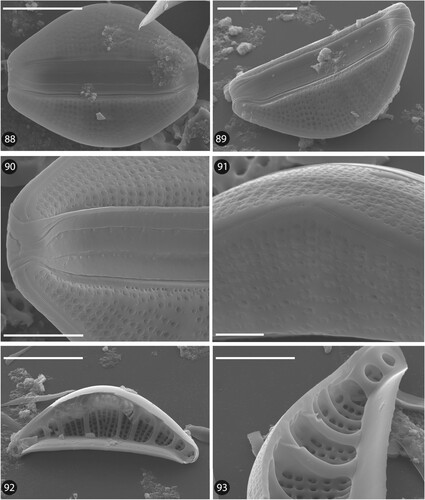
Fig. 94. Phylogenetic position Gomphonema bourbonense (ASV11 and ASV16) in the ML tree. Bootstrap values are given for each node and the scale bar gives the number of substitutions per site. ‘*’ indicates sequences added in the phylogeny using the multifurcating constraint.

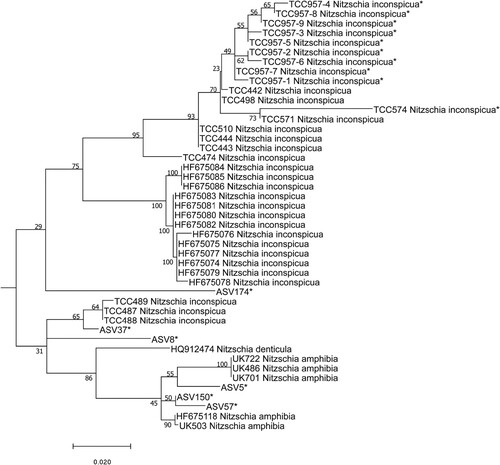
Fig. 95. Phylogenetic position of undetermined ASV (ASV5, ASV8, ASV37, ASV57, ASV150) of the Nitzchia complex in the ML tree. Bootstrap values are given for each node and the scale bar gives the number of substitutions per site. ‘*’ indicates sequences added in the phylogeny using the multifurcating constraint.
Undetermined sequence ASV22 was also identified to family level, namely Naviculaceae, with a bootstrap value of 99% using DADA2. BLAST identified ASV22 as Nupela Vyverman & Compère sp. with 93% identity. Although the bootstrap value (45%) only gave poor support for its position in the phylogeny, this sequence was next to a sequence of Nupela (Fig. 12). We therefore correlated the ASV22 read number to the frustule counts of Nupela sp., and the results were significant (R2 = 0.58, p < 0.05). We therefore concluded that this sequence belongs to a species of Nupela. LM observations (Figs 13–53) and SEM observations (Figs 54–59) confirmed the genus affiliation. According to the molecular, LM and SEM observations, this species is new to science and is described below (Taxonomic results).
Undetermined sequence ASV38 was assigned to the genus Epithemia Kützing using DADA2. BLAST identified ASV38 with 95% identity to four sequences belonging to Epithemia gibba (Ehrenberg) Kützing, Epithemia hyndmanii W. Smith and Epithemia sp. The position of ASV38 in the phylogeny was inside the Epithemia clade (Fig. 60). Earlier identifications carried out for the iconographic atlas of the French West Indies identified a taxon as Rhopalodia sp.1 (Eulin et al. Citation2017e). However, this species is now part of Epithemia since Rhopalodia O. Muller has been merged with Epithemia on the basis of morphological and genetic data (Ruck et al. Citation2016). LM (Figs 61–87) and SEM observations (Figs 88–93) of our Epithemia sp. present morphological features which differ from closely related known species. ASV38 reads number and frustule counts of Epithemia sp. were significantly correlated (R2 = 0,1223, p < 0.05). Based on the molecular, LM and SEM results, this species is new to science and is described below (Taxonomic results).
Intermediate case: DADA2 assigned sequence ASV11 to the genus Gomphonema Ehrenberg, with 100% bootstrap support. BLAST identified the sequence as G. bourbonense E. Reichardt with 97% identity. The position of ASV11 in the phylogenetic tree (Fig. 94) shows that it belongs to the G. bourbonense clade, which also includes ASV16. The LM determinations carried out in the earlier study (Eulin et al. Citation2017b) show that two morphologically similar species were identified in the West Indies: G. bourbonense and G. designatum E. Reichardt. However, we suspected that these identifications were erroneous. We therefore carried out detailed morphometric analyses on the West Indies samples in which these species were formerly identified. Our measurements, given in Table , did not fit with G. designatum (particularly the ranges for length, width and stria density) but with G. bourbonense. Therefore we considered the former determinations of G. designatum by Eulin et al. (Citation2017b) should be G. bourbonense. Correlation between the numbers of reads of ASV11 plus ASV16 and frustule counts of G. bourbonense (to which were added frustule counts previously identified as G. designatum) was significant. Therefore, although two species were identified and illustrated in the atlas of these islands (Eulin et al. Citation2017b), we conclude that they constitute a single species, G. bourbonense.
Table 1. Morphological comparison between Gomphonema bourbonense and Gomphonema designatum.
Complex case: A group of sequences were treated together due to their close phylogenetic affiliation. These were ASV5, ASV8, ASV37, ASV57 and ASV150. DADA2 assigned ASV5, ASV8, ASV37, ASV57 and ASV150 to the genus Nitzschia Hassall. BLAST gave the following identifications: Nitzschia amphibia Grunow (98%) (ASV5), Nitzschia inconspicua Grunow (97%) (ASV8), N.inconspicua (98%) (ASV37), N. amphibia (98%) (ASV57) and N. amphibia (98%) (ASV150). The position of these five sequences is shown in the phylogenetic tree (Fig. 95); their positions next to N. inconspicua and N. amphibia are loosely supported. Therefore, all these sequences (ASV5, ASV8, ASV37, ASV57, ASV150) were considered for the correlation between their read numbers and the frustule counts of several Nitzschia species. Given their position in the phylogeny, we suspected that they might correspond to N. inconspicua, N. amphibia, Nitzschia denticula Grunow, but also to other morphologically similar taxa illustrated in Eulin et al. (Citation2017e) as N. frustulum (Kützing) Grunow; N. frustulum forme 2, N. frustulum forme 3, N. sp. 64 and N. sp. 41. The correlations (supplementary data 4) show that in several cases, read abundance of an ASV could be correlated to several species abundances identified by LM (e.g. ASV5 correlated with N. inconspicua and N. frustulum, ASV8 correlated with N. inconspicua and N. amphibia). Moreover, some ASVs co-occurred (e.g. ASV5 and ASV8), although they were not located in the same phylogenetic clade (ASV5 in N. amphibia clade, ASV8 weakly placed in the phylogeny between N. amphibia, N. inconspicua, N. denticula) and were correlated with the same species identified in LM (N. inconspicua). For all these reasons, it is impossible to give species names to these ASVs with any certainty.
Taxonomic results
Nupela boucheziae Kochoska, Chardon, Chonova, Keck, Kermarrec, Larras, S.F. Rivera, Tapolczai, Vasselon, Levkov & Rimet sp. nov. (Figs 13–59)
Description: LM (Figs 13–53): Frustules convex, heterovalvar, slightly asymmetric about the apical plane. Valves lanceolate, elliptical-lanceolate with slightly rounded to sub-rostrate apices, 7.0–15.0 µm long and 4.0–4.5 µm wide. One valve with long raphe slits, thread-like and almost straight, incomplete on the other valve, reduced to a small helictoglossa, ‘ghost’ full raphe. Axial area linear and narrow in both valvae, central area small to very small, round to elliptical. Striae and areolae not visible in LM.
Description: SEM (Figs 54–59): Heterovalvar frustules. Proximal raphe ends externally expanded and internally simple (Figs 55, 58). Terminal raphe ends curved externally to the same side of the valve and internally ending in small helictoglossae (Figs 54, 56 and 58, 59). Reduced raphe valve with a smooth axial area without depressions (Figs 57, 59). Transapical striae are slightly radial to parallel towards the apices, 30–40 in 10 µm, composed of continuous lines of areolae, ca. 50 in 10 µm. Outer openings of areolae are round and occluded by a delicate hymen. Inner openings of areolae larger than the outer, round to oval.
Type: France, Blanche-Pont de l'Alma in Martinique, biofilm, collection date: 12.05.2018; Leg. Anne Eulin, Estelle Lefrançois; Coordinates: −61.08895606 latitude, 14.70644106 longitude.
Holotype slide and treated material: Accession No. PC0643142 (Museum National d’Histoire Naturelle, Paris, France).
Isotype slide and treated material: Accession No. MKNDC 14432 (Institute of Biology, Skopje, Republic of North Macedonia). Slide and treated material TCC1086 (Thonon Culture Collection).
Etymology: This species is dedicated to Dr. Agnès Bouchez, who made major contributions to the development of metabarcoding and diatom science.
Taxonomic remarks: Based on the valve shape, N. boucheziae is comparable to several Nupela species (see Table ). Nupela boucheziae is similar to N. praecipuoides Tremarin & T. Ludwig (Tremarin et al. Citation2015) but N. praecipuoides has one valve with long raphe slits while the other valve is araphid. Clear differences between N. boucheziae and N. praecipuoides can also be seen in that the axial area is linear and narrow on the raphid valve, but lanceolate, smooth or generally with irregular depressions on the araphid valve that may or not be visible in LM. Nupela praecipuoides also has much lower areola and stria densities. Specimens similar to N. praecipuoides were recorded by Rumrich et al. (Citation2000) as N. spec. cf. praecipua in Equador, and in south Brazil as N. praecipua (E. Reichardt) E. Reichardt by Schneck et al. (Citation2008), Tremarin et al. (Citation2009) and Moresco et al. (Citation2011). Nupela praecipuoides was also recorded in rivers of the Atlantic forest in southern Brazil (Tremarin et al. Citation2015).
Table 2. Morphological features and measurements of Nupela boucheziae sp. nov. and similar species. n.a.: no data available.
Nupela boucheziae is similar to N. praecipua described from Mexico (Reichardt Citation1988). Both species share a similar valve outline, but the striae and areolae of N. praecipua are coarser (32–36 striae in 10 µm, 30–35 areolae in 10 µm) than those of N. boucheziae: this is easily seen under LM. Furthermore, N. praecipua has smaller valves (length: 8.0–13.5 µm) and deep depressions in the axial area of the araphid valve, which is not the case for N. boucheziae. Nupela praecipua has slightly convergent striae at the apices, unlike N. boucheziae (Reichardt Citation1988, Rumrich et al. Citation2000). Some similarity was observed between N. boucheziae and N. chilensis (Krasske) Lange–Bertalot (Lange–Bertalot et al. Citation1996) in valve outline and striation pattern. However, N. chilensis has larger valves (length: 16–26, width: 5–7 µm), a wider central area, long raphe slits on both valves, and lower stria density (30–32 in 10 µm) than N. boucheziae. Nupela difficilis Straube, Tremarin & T. Ludwig is mainly characterized by its lanceolate valve outline, subrostrate apices, and asymmetric central area, as well as the straight, interior, proximal raphe ends (Tremarin et al. Citation2015). Nupela difficilis has a similar valve outline and size to N. boucheziae. However there are clear differences with a raphe on both valves and strongly convex valve margins with its asymmetric central area, reaching the valve margin on one side.
Nupela decipiens (Reimer) Potapova is comparable in size and stria density to N. boucheziae (Potapova Citation2013). Nupela decipiens can be differentiated by the narrowly rostrate to subrostrate apices, the size and shape of the central area (in raphid valve widely rounded, in araphid valve widely lanceolate) not reaching the valve margin. The axial area also differs between the two species: lanceolate to widely lanceolate, with irregular external depressions, the longitudinal depression in the araphid valve resembling a raphe under LM. Nupela neglecta Ponader, Lowe & Potapova, and N. boucheziae share similar valve outlines (Potapova et al. Citation2003); however, N. neglecta has slightly protracted apices and quite different valve margins: one valve is convex to parallel in the middle, the other slightly concave and often slightly asymmetrical about apical and transapical planes. Nupela neglecta has raphe slits on both valves (one raphe shorter than the other), and a higher stria density (40–48 in 10 µm) compared to N. boucheziae. Nupela jahniae–reginae Lange–Bertalot & Metzeltin (Rumrich et al. Citation2000) also has a comparable valve shape to N. boucheziae. (elliptical lanceolate to lanceolate). But there are differences between the two species: N. jahniae–reginae has obtuse, slightly protracted apices, convex to slightly parallel in the middle valve margins, with a longer raphe on the short raphe valve and lower valve width (3.0–4.0 µm in N. jahniae–reginae) and higher stria density (ca. 50 in 10 µm in N. jahniae–reginae).
Ecological remarks: This species is abundant in the West Indies (Martinique and Guadeloupe). The type locality is situated in the upstream stretch of the Alma river. It is characteristic of low nutrients and organic matter (Eulin et al. Citation2017d).
Slides are deposited at the National Museum of Natural History (MNHN) in Paris, France and the Macedonian National Diatom Collection (MKNDC) at the Institute of Biology, Faculty of Natural Sciences, Skopje, Republic of North Macedonia.
Epithemia boucheziae Kochoska, Chardon, Chonova, Keck, Kermarrec, Larras, S.F. Rivera, Tapolczai, Vasselon, Levkov & Rimet sp. nov. (Figs 61–93)
Description. LM (Figs 61–87): Frustules lanceolate to linear-elliptic in girdle view. Valves semi-elliptical to almost triangular, dorsal edge of valve convex, ventral edge slightly concave. The valve ends slightly protracted and curved towards the ventral edge of the valve. Valves 17.0–23.0 µm long and 6.5–8.0 µm in width. The raphe is located in a channel along the dorsal edge of the valve. Areolae visible with LM, ca. 16 in 10 µm.
Description. SEM (Figs 88–93): Externally, proximal raphe ends almost straight, slightly expanded to droplet shaped. Terminal raphe ends curved ventrally. Internal raphe endings are simple. Raphe opens internally into a canal with small round holes (portulae) lying between the major transapical ribs (Fig. 93). Striae coarsely punctate and strongly radiate. Externally, striae are composed of complex areolae: near the ventral side these are composed of two opposed ‘C’ shaped slits, and near the dorsal side of four opposed ‘C’ shaped slits, giving a flower-like aspect (Fig. 91). Discontinuous stria pattern with several missing areolae in the middle part of the valve (Fig. 89). Striae uniseriate, 16–18 in 10 µm. Primary fibular costae 3–4 in 10 µm with usually 4–6 striae between two costae (Figs 92, 93).
Type. France: Rivière Bras David in Guadeloupe, biofilm, collection date: 16.04.2019; Leg. Anne Eulin, Estelle Lefrançois; Coordinates: −61.67076118 latitude, 16.19470576 longitude.
Holotype slide and treated material: Accession No. PC0643143 (Museum National d’Histoire Naturelle, Paris, France).
Isotype slide and treated material: Accession No. MKNDC 14433 (Institute of Biology, Skopje, Republic of North Macedonia). Slide and treated material TCC1088 (Thonon Culture Collection).
Etymology:This species is dedicated to Dr. Agnès Bouchez, on behalf of her close colleagues and former students who all enjoyed working with her.
Taxonomic remarks: Based on morphological features, E. boucheziae is similar to several species previously placed in Rhopalodia (Table ). Valve shape of E. boucheziae is similar to R. michelorum Krammer, which has a nearly straight ventral margin, and quite distinct ends, which are narrowly protracted, frequently capitate, and bent ventrally (Krammer Citation1988). However, R. michelorum has bigger valves (length: 17–50 µm, width: 6–10 µm), its proximal raphe ends are curved to the same side, hook-like (E. boucheziae has straight proximal raphe ends), and the striae are composed of single rows of areolae. The stria pattern is discontinuous with several missing areolae in the middle part of the valve. Stria density is higher in R. michelorum (19–24 in 10 µm) than E. boucheziae (16–18 in 10 µm) and the primary fibular costae of R. michelorum are very stout and more distantly spaced (1.5–2.8 in 10 µm, Table ).
Table 3. Morphological features and measurements of Epithemia boucheziae sp. nov. and similar species. n.a.: no data available.
Epithemia boucheziae is usually smaller than Rhopalodia gibberula (Ehrenberg) O. Müller, however, the smaller individuals of R. gibberula are similar to E. boucheziae. The difference can be seen in the valve outline, which is linear-elliptic to broad-elliptical in R. gibberula and hardly or not retracted in the middle, and the sides are massiform to strongly convex. Frustules of R. gibberula are sickle shaped, the dorsal side strongly convex, the ventral side strongly concave in large forms, almost parallel to the dorsal side, in smaller ones the ventral side is less curved. The difference between both species is easily noted in the proximal raphe ends: R. gibberula has a fissure in the central portion and areolae with tube-like processes and bordered by C-shaped foramina (Lange–Bertalot & Krammer Citation1987). A population of R. gibberula was recorded from freshwater assemblages in Guadeloupe by Bourrelly & Manguin (Citation1952) (Table , ref. 5), but this has bigger valves, higher stria density, and a different valve outline. Bourrelly & Manguin (Citation1952) observed different varieties of the ‘gibberula’ group in freshwaters of Guadeloupe: R. gibberula var. miniuens O. Müller, R. gibberula var. succincta (Brébisson) Fricke, R. gibberula var. vanheurckii O. Müller and several others. Rhopalodia gibberula var. miniuens can be differentiated by its elliptical, semicircular valve outline, the apices are protracted and bent ventrally and the dorsal side is strongly convex, but the ventral side is straight. This species is also wider, with higher costa and stria densities. Rhopalodia gibberula var. succincta has similarities with E. boucheziae in valve shape and apices, but it is smaller and has higher costa and stria densities. Epithemia boucheziae is comparable to R. gibberula var. vanheurckii but differences are clear in the valve outline (linear-elliptical, retracted in the middle in R. gibberula var. vanheurckii), and R. gibberula var. vanheurckii is narrower, has more prolonged apices, a larger frustule and a higher stria density (Table ).
Epithemia musculus Kützing is similar to E. boucheziae, characterized by broadly elliptical frustules in girdle view, usually with rounded apices (Krammer Citation1988). Valves have strongly convex dorsal margins and straight ventral margins, apices are bent ventrally and rounded. This valve outline is different from that of E. boucheziae. Epithemia musculus is also larger, with more densely arranged, uniseriate striae comprising contrastingly structured areolae. The latter are stout, large and very distinct, less than 15/10 µm, with multiple lips in the foramina, which are unique to E. musculus (Table ).
Rhopalodia acuminata Krammer is morphologically similar to E. boucheziae. Its most specific differences are in its shape: the frustule is sickle-shaped, with a strongly convex dorsal margin and weakly concave ventral margin. The areolae in R. acuminata are uniseriately arranged with some double rows beside the raphe canal. Areolae are occluded externally by circular or C-shaped slits in E. boucheziae. Rhopalodia acuminata is also larger than E. boucheziae.
Rhopalodia brebissonii Krammer also resembles E. boucheziae. This species is characterized by broadly elliptical frustules and valves with a strongly convex dorsal margin, straight or slightly concave ventral margin, and protracted and ventrally bent apices (Krammer Citation1988). The main differences between R. brebissonii and E. boucheziae are in their stria structure, with striae in R. brebissonii composed of double rows of areolae on both sides of the raphe canal and a single row on the rest of the valve, while E. boucheziae has striae composed of complex areolae. In addition, R. brebissonii has larger valves and higher stria density (17–22 in 10 µm) compared to E. boucheziae.
Slides are deposited at the National Museum of Natural History in Paris (MNHN) France and the Macedonian National Diatom Collection (MKNDC) at the Institute of Biology, Faculty of Natural Sciences, Skopje, Republic of North Macedonia.
Discussion
Rimet et al. (Citation2018) proposed a methodology to use HTS sequencing data of environmental samples to define barcodes for species which have no sequence in reference barcoding libraries. When we applied this approach to our samples from the West Indies (Guadeloupe, Martinique), the applicability of this approach was variable. We distinguished three categories depending on the level of support between the criteria used in this approach, from simple to complex.
Simple cases: The first group is a category for which correspondence between an unidentified sequence and a morphological specimen can easily be established. These cases were simple because no closely related species were present in the samples. In some cases, the morphological form could be easily assigned to an existing species (U. goulardii), in the other cases it concerned species new to science (N. boucheziae, E. boucheziae).
The first case (ASV2) concerned a sequence that had the highest number of ASV reads in the rivers of the West Indies. However, it was only identified to the family level (Fragilariaceae) with Diat.barcode version 9 because the latter did not contain any similar reference barcode to allow correct naming. Based on molecular and LM data, we could identify this sequence as U. goulardii, a species recently transferred from Fragilaria to Ulnaria (Wetzel et al. Citation2022). No similar or related species were observed with LM or in the sequencing data, which made this case straightforward.
Another case was a sequence (ASV22) assigned to the genus Nupela on the basis of molecular data. As LM and SEM observations did not match any of the most morphologically similar described species in this genus (Potapova et al. Citation2003, Potapova Citation2011, Tremarin et al. Citation2015), this taxon was described as a new species, N. boucheziae.
The last simple case concerned another new species belonging to Epithemia. This taxon had previously been observed in the West Indies islands and referred to as Rhopalodia sp1 by Eulin et al. (Citation2017e). Recently Rhopalodia was merged with Epithemia (Ruck et al. Citation2016). The new species, E. boucheziae, is an a example of a new taxon established using several criteria, following the prerequisites of the integrative taxonomy concept.
Intermediate cases: The second group comprises cases where there is also a relatively easy correspondence between genetic and morphological criteria. However, these cases are more complex because morphologically similar species have been described in the literature, which can lead to misidentifications. Earlier LM observations carried out to establish the regional Atlas of the West Indies (Eulin et al. Citation2017b) identified two morphologically similar species, G. bourbonense and G. designatum. Careful morphological re-examination of the samples in the present study demonstrated that all the specimens belonged to G. bourbonense, as several morphometric features were outside the ranges for G. designatum. In addition, the two sequences (ASV11, ASV16) were obtained from the same samples and both were placed in the G. bourbonense clade. We therefore concluded that the species in our samples was G. bourbonense.
Complex case: The last category concerns groups of several, genetically similar sequences and morphologically similar forms that were impossible to resolve. Three frequent (ASV5, ASV8 and ASV37) and two rarer (ASV57, ASV150) sequences, all belonging to the genus Nitzschia, were in the same clade (N. inconspicua, N. amphibia). Based on the morphology of the specimens present in the samples in which these sequences were frequent, they could match several species (N. inconspicua, N. amphibia, N. denticula and N. frustulum), but also morphologically similar forms previously referred to as N. frustulum forma 2, N. frustulum forma 3, Nitzschia sp. 64 and Nitzschia sp. 41 (Eulin et al. Citation2017e). In several cases, the read abundance of a single ASV was correlated with several species abundances identified by LM. In addition some ASVs that were not part of the same clade, co-occurred together and were correlated with the same species in LM. There are several possible explanations for this lack of correspondence. For instance, the correspondence between genetic and morphological criteria may sometimes exist only for morphological features which are usually not taken into account for species discrimination (e.g. Gomphonema parvulum [Kützing] Kützing in Kermarrec et al. Citation2013). Another explanation is that some morphological species are composed of several cryptic species (and genotypes) that do not co-occur, which hampers the correspondence between morphological and genetic criteria (Trobajo et al. Citation2009). This example clearly shows the limits of Rimet et al. (Citation2018) method.
These West Indies islands host more than 100 morphologically identified species. We could establish a clear link with one or several barcodes in Diat.barcode v.10 for seven species, of which four species are illustrated here. Five were simple cases (U. goulardii, N. boucheziae, E. boucheziae, Sellaphora nigri (De Notaris) C.E. Wetzel & L. Ector, Navicula incarum U. Rumrich & Lange-Bertalot), and two were intermediate cases (G. bourbonense/designatum, Navicula escambia/symmetrica/simulata). These were all essentially abundant species. For rarer species, the method could not be applied. Cell isolation and culturing followed by an integrative taxonomical approach is the only solution to resolving such problematic taxonomic groups.
Conclusion
Metabarcoding is a promising approach that simplifies diatom identification and overcomes the problems associated with the traditional morphological approach (Vasselon et al. Citation2019), however, diatoms exhibit high diversity (e.g. Levkov et al. Citation2007, Mann & Vanormelingen, Citation2013) and strong endemism (Chonova et al. Citation2021, Verleyen et al. Citation2021, Rimet et al. Citation2023). To correctly identify species and use them effectively for assessing ecological quality, it is necessary to continue exploring their diversity, especially in poorly studied areas, such as the tropics. Moreover, to describe species and accurately define their boundaries, it is necessary to use additional criteria to the morphological criteria, such as molecular criteria (Dayrat Citation2005).
Work to expand the Diat.barcode library of the West Indies using Rimet et al. (Citation2018) methodology now allows the majority of diatom environmental sequences in rivers to be identified. The proportion of sequences identified to species level has been increased from 45% to 84% in the latest version of Diat.barcode (v.10) with all necessary metadata (Rimet et al. Citation2021a, Citationb). Furthermore, our newly described species can easily be identified even in the presence of morphologically sister species (e.g. Evans et al. Citation2009, Rivera et al. Citation2018a, Citationb) since they have associated barcodes. However, even if Rimet et al. (Citation2018) method enables quick and cost-effective completion of the reference barcoding library, in some cases it is impossible to apply to a complex taxonomic group, especially when several similar taxa are present in the microscope samples and amplicon data. In this case, we recommend using isolation and culturing methods alongside a careful morphological study.
Supplementary data 4. Spearman’s correlation between frustule counts of suspected species and percentage of reads of ASVs. (Correlation coefficient: bottom left of the table, p-value: upper right of the table.)
Download MS Word (20.2 KB)Supplementary data 3. SEM micrographs of Epithemia boucheziae sp. nov. Figs 1–2. External valve view. Figs. 3–4 Internal valve view. Scale bars = 10 μm.
Download JPEG Image (426.4 KB)Supplementary data 2. LM micrographs of Epithemia boucheziae sp. nov. Figs 1–10. Valve view. Figs 11–13c. Girdle view, from river Rivière Bras David in Guadeloupe. Scale bar = 10 μm.
Download JPEG Image (532.9 KB)Supplementary data 1. LMmicrographs of Nupela boucheziae sp. nov. Figs 1–23. Valves with raphe and Figs. 1–19. Valves without raphe, from a site called Blanche-Pont de l’Alma in Martinique. Scale bar = 10 μm.
Download JPEG Image (317.4 KB)Acknowledgments
The authors gratefully acknowledge Anne Eulin-Garrigue for providing the results of her previous work on the same sites. Zlatko Levkov received support from the Alexander von Humboldt Foundation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary data
Supplemental data for this article can be accessed at https://doi.org/10.1080/0269249X.2023.2237977. Anne Eulin-Garrigue and Estelle Lefrancois provided the supplemental data when floristic counts were made in the framework of the monitoring of rivers (Eulin et al. Citation2017a, Citationb, Citationc, Citationd, Citatione, Citationf). Figure legends for Supplementary data: Supplementary data 1. LM micrographs of Nupela boucheziae sp. nov. Figs 1–23. Valves with raphe and Figs. 1–19. Valves without raphe, from a site called Blanche-Pont de l'Alma in Martinique. Scale bar = 10 m. Supplementary data 2. LM micrographs of Epithemia boucheziae sp. nov. Figs 1–10. Valve view. Figs 11–13c. Girdle view, from river Riviere Bras David in Guadeloupe. Scale bar = 10 m. Supplementary data 3. SEM micrographs of Epithemia boucheziae sp. nov. Figs 1–2. External valve view. Figs. 3–4 Internal valve view. Scale bars = 10 m. Supplementary data 4. Spearman's correlation between frustule counts of suspected species and percentage of reads of ASVs. (Correlation coefficient: bottom left of the table, p-value: upper right of the table.)
Additional information
Funding
References
- Abarca N., Jahn R., Zimmermann J., & Enke N. 2014. Does the cosmopolitan diatom Gomphonema parvulum (Kutzing) Kutzing have a biogeography? PLoS ONE 9: 1–18. http://doi.org/10.1371/journal.pone.0086885
- Afnor. 2003. Norme française NF EN 13946. Qualite de l’eau - Guide Pour l’echantillonnage En Routine et Le Pretraitement Des Diatomees Benthiques de Rivieres., 1–18.
- Afnor. 2016. Norme française NF T90–354 Avril 2016. Qualité de l’eau – Échantillonnage, traitement et analyse de diatomées benthiques en cours d’eau et canaux. La Plaine Saint-Denis Cedex: Association Française de Normalisation (AFNOR); p. 1–119.
- Altschul F.S., Gish W., Miller W., Myers W.E. & Lipman J.D. 1990. Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. http://doi.org/10.1016/S0022-2836(05)80360-2.
- Barbour M.T., Gerritsen J., Snyder B.D. & Stribling J.B. 1999. Rapid bioassessment protocols for use in streams and wadeable rivers: periphyton, benthic macroinvertebrates, and fish. Second edition. – EPA 841–B–99–002. US Environmental Protection Agency, Office of Water, Washington, DC.
- Bourrelly P. & Manguin E. 1952. Algues d'eau douce de la Guadeloupe et dépendances. Centre National de la Recherche Scientifique, Société d'Edition d'Enseignement Supérieur, Paris: 98–100.
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A. & Holmes S.P. 2016. DADA2: high high-resolution sample inference from illumina amplicon data. Nature Methods 13: 581–583. http://doi.org/10.1038/nmeth.3869.
- Canino A., Bouchez A., Laplace-Treyture C., Domaizon I. & Rimet F. 2021. Phytool, a ShinyApp to homogenise taxonomy of freshwater microalgae from DNA barcodes and microscopic observations. Metabarcoding and Metagenomics 5: 199–205. http://doi.org/10.3897/mbmg.5.74096
- Cen. 2014. EN 13946. Water quality—Guidance standard for the routine sampling and pretreatment of benthic diatoms from rivers. CEN-CEN ELEC, 1–17.
- Cen. 2018. Water quality – CEN/TR 17245 – Technical report for the routine sampling of benthic diatoms from rivers and lakes adapted for metabarcoding analyses. CEN standard. CEN-CENELEC Management Centre: Rue de la Science 23, B-1040 Brussels, pages 1–8.
- Chonova T, Rimet F, Bouchez A, & Keck F. 2021. Revisiting global biogeography of freshwater diatoms: new insights from molecular data. ARPHA Conference Abstracts 4: e65129. http://doi.org/10.3897/aca.4.e65129
- Dayrat B. 2005. Towards integrative taxonomy. Biological Journal of the Linnean Society 85: 407–415. http://doi.org/10.1111/j.1095-8312.2005.00503.x
- Ehrenberg C.G. 1843. Verbreitung und Einfluss des mikroskopischen Lebens in Süd-und Nord-Amerika. Abhandlungen der Königlichen Akademie der Wissenschaften zu Berlin: 291–445, 4 pls.
- Eulin A., Lefrançois E., Delmas F., Coste M., Gueguen J. & Rosebery J. 2017a. Flore des diatomées des Antilles françaises. Volume 1. Agence française pour la biodiversité, Office de l’eau Guadeloupe, Office de l’eau Martinique, Irstea, DEAL Martinique, DEAL Guadeloupe. 145 pp.
- Eulin A., Lefrançois E., Delmas F., Coste M., Gueguen J. & Rosebery J. 2017b. Flore des diatomées des Antilles françaises. Volume 2. Agence française pour la biodiversité, Office de l’eau Guadeloupe, Office de l’eau Martinique, Irstea, DEAL Martinique, DEAL Guadeloupe. 147 pp.
- Eulin A., Lefrançois E., Delmas F., Coste M., Gueguen J., & Rosebery J. 2017c. Flore des diatomées des Antilles françaises. Volume 3. Agence française pour la biodiversité, Office de l’eau Guadeloupe, Office de l’eau Martinique, Irstea, DEAL Martinique, DEAL Guadeloupe. 132 pp.
- Eulin A., Lefrançois E., Delmas F., Coste M., Gueguen J. & Rosebery J. 2017d. Flore des diatomées des Antilles françaises. Volume 4. Agence française pour la biodiversité, Office de l’eau Guadeloupe, Office de l’eau Martinique, Irstea, DEAL Martinique, DEAL Guadeloupe. 133 pp.
- Eulin A., Lefrançois E., Delmas F., Coste M., Gueguen J. & Rosebery J. 2017e. Flore des diatomées des Antilles françaises. Volume 5. Agence française pour la biodiversité, Office de l’eau Guadeloupe, Office de l’eau Martinique, Irstea, DEAL Martinique, DEAL Guadeloupe. 170 pp.
- Eulin A., Lefrançois E., Delmas F., Coste M., Gueguen J. & Rosebery J. 2017f. Flore des diatomées des Antilles françaises. Volume introductif. Agence française pour la biodiversité, Office de l’eau Guadeloupe, Office de l’eau Martinique, Irstea, DEAL Martinique, DEAL Guadeloupe. 44 pp.
- European Commission 2000. Directive 2000/60/EC of the European parliament and of the council of 23rd October 2000 establishing a framework for community action in the field of water policy. Official Journal of the European Communities 327: 1–72.
- European Committee for Standardisation 2014a. EN 13946 – Water quality – Guidance for the routine sampling and preparation of benthic diatoms from rivers and lakes. – 18 pp., Afnor, La Plaine St Denis, France.
- European Committee for Standardisation 2014b. EN 14407 – Water quality – Guidance for the identification and enumeration of benthic diatom samples from rivers and lakes. – 13 pp., Afnor, La Plaine St Denis, France.
- Evans K.M., Chepurnov V.A., Sluiman H.J., Thomas S.J., Spears B.M. & Mann D.G. 2009. Highly differentiated populations of the freshwater diatom Sellaphora capitata suggest limited dispersal and opportunities for allopatric speciation. Protist 160: 386-396. http://doi.org/10.1016/j.protis.2009.02.001
- Evans K.M., Wortley A.H. & Mann D.G. 2007. An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 158: 349–364. http://doi.org/10.1016/j.protis.2007.04.001
- Gomez F., Lopez–Garcia P., Dolan J.R. & Moreira D. 2012. Molecular phylogeny of the marine dinoflagellate genus Heterodinium (Dinophyceae). European Journal of Phycology 47: 95–104. http://doi.org/10.1080/09670262.2012.662722
- Guillou L., Bachar D., Audic S., Bass D., Berney C. & Bittner L. 2013. The protist ribosomal reference database (PR2): A catalog of unicellular eukaryote small sub-unit rrna sequences with curated taxonomy. Nucleic Acids Research 41: D597–D604. http://doi.org/10.1093/nar/gks1160
- Hall T.A. 1999. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/Nt. Nucleic Acids Symposium Series 41: 95-98.
- Hamilton P. B., Lefebvre K. E. & Bull R. D. 2015. Single cell PCR amplification of diatoms using fresh and preserved samples. Frontiers in Microbiology 6: 1084. http://doi.org/10.3389/fmicb.2015.01084
- Hamilton P. B., Savoie A. M., Sayre C. M., Skibbe O., Zimmermann J. & Bull R. D. 2019. Novel neidium Pfizer species from western Canada based upon morphology and plastid DNA sequences. Phytotaxa 419: 39-62.
- Hausmann S., Charles D. F., Gerritsen J. & Belton T. J. 2016. A diatom-based biological condition gradient (BCG) approach for assessing impairment and developing nutrient criteria for streams. Science of the Total Environment 562: 914–927. http://doi.org/10.1016/j.scitotenv.2016.03.173
- Hebert P. D. & Gregory T. R. 2005. The promise of DNA barcoding for taxonomy. Systematic Biology 54: 852–859. http://doi.org/10.1080/10635150500354886
- Jaramillo A., Osman, D., Caputo, L., & Cardenas, L. 2015. Molecular evidence of a Didymosphenia geminata (Bacillariophyceae) invasion In Chilean freshwater systems. Harmful Algae 49: 117–123. http://doi.org/10.1016/j.hal.2015.09.004
- Kahlert M., Albert R. L., Anttila E. L., Bengtsson R., Bigler C., Eskola T., Galman V., Gottschalk S., Herlitz E., Jarlman A., Kasperoviciene J., Kokocinski M., Luup H., Miettinen J., Paunksnyte I., Piirsoo K., Quintana I., Raunio J., Sandell B., Simola H., Sundberg I., Vilbaste S. & Weckstrom J. 2009. Harmonization is more important than experience-results of the first Nordic–Baltic diatom intercalibration exercise 2007 (stream monitoring). Journal of Applied Phycology 21: 471–482. http://doi.org/10.1007/s10811-008-9394-5
- Keck F., Vasselon V., Rimet F., Bouchez A. & Kahlert M. 2018. Boosting DNA metabarcoding for biomonitoring with phylogenetic estimation of operational taxonomic units’ ecological profiles. Molecular Ecology Resources 18(6): 1299–1309. http://doi.org/10.1111/1755-0998.12919
- Kelly M. G., Trobajo R., Rovira L. & Mann D. G. 2015. Characterizing the niches of two very similar Nitzschia species and implications for ecological assessment. Diatom Research 30: 27–33. http://doi.org/10.1080/0269249X.2014.951398
- Kelly M., Urbanic G., Acs E., Bennion H., Bertrin V., Burgess A., Denys L., Gottschalk S., Kahlert M., Karjalainen S. M., Kennedy B., Kosi G., Marchetto A., Morin S., Picinska-Fałtynowicz J., Poikane S., Rosebery J., Schoenfelder I., Schoenfelder J. & Varbiro G. 2014. Comparing aspirations: intercalibration of ecological status concepts across European lakes for littoral diatoms. Hydrobiologia 734: 125–141. http://doi.org/10.1007/s10750-014-1874-9
- Kermarrec L., Bouchez A., Rimet F. & Humbert J. F. 2012. First evidence of the existence of semi-cryptic species and of a phylogeographic structure in the Gomphonema parvulum (Kutzing) Kutzing complex (Bacillariophyta). Protist 164: 686–705. http://doi.org/10.1016/j.protis.2013.07.005
- Kermarrec L., Franc A., Rimet F., Chaumeil P., Humbert J. F. & Bouchez A. 2013. Next-generation sequencing to inventory taxonomic diversity in eukaryotic communities: a test for freshwater diatoms. Molecular Ecology Resources 13: 607–619. http://doi.org/10.1111/1755-0998.12105
- Kermarrec L., Franc A., Rimet F., Chaumeil P., Frigerio J. M., Humbert J. F. & Bouchez A. 2014. A next-generation sequencing approach to river biomonitoring using benthic diatoms. Freshwater Science 33: 349–363. http://doi.org/10.1086/675079
- Khan–Bureau D. A., Morales E. A., Ector L., Beauchene M. S. & Lewis L. A. 2016. Characterization of a new species in the genus Didymosphenia and of Cymbella janischii (Bacillariophyta) from Connecticut, USA. European Journal of Phycology 51(2): 203–216. http://doi.org/10.1080/09670262.2015.1126361
- Krammer, K. 1988. The gibberula-group in the genus Rhopalodia O. Müller (Bacillariophyceae). II. Revision of the group and new taxa. Nova Hedwigia 47: 159–205.
- Krasske, G. 1939. Zur Kieselalgenflora Südchiles. Archiv für hydrobiologie 35: 349–468.
- Kumar S., Stecher G. & Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology And Evolution 33(7): 1870–1874. http://doi.org/10.1093/molbev/msw054
- Lange–Bertalot H., Külbs K., Lauser T., Nörpel–Schempp M. & Willmann M. 1996. Diatom taxa introduced by Georg Krasske. Documentation and revision. Dokumentation und Revision der von Georg Krasske beschriebenen Diatomeen-Taxa. Iconographia Diatomologica 3: 1–358.
- Lange–Bertalot H. & Krammer K. 1987. Bacillariaceae, Epithemiaceae, Surirellaceae. Neue und wenig bekannte Taxa, neue Kombinationen und Synonyme sowie Bemerkungen und Ergänzungen zu den Naviculaceae. Bibliotheca Diatomologica 1: 1–289.
- Lange-Bertalot H. & Moser G. 1994. Brachysira. Monographie der Gattung. Bibliotheca Diatomologica 29: 1–212.
- Lefebvre K. E., Hamilton P. B. & Pick F. R. 2017. A comparison of molecular markers and morphology for Neidium taxa (Bacillariophyta) from Eastern North America. Journal of Phycology 53: 680–702.
- Levkov Z., Krstic S., Metzeltin D. & Nakov T. 2007. Diatoms of lakes Prespa and Ohrid (Macedonia). Iconographia Diatomologica 16: 1–603.
- Mann D.G. & Vanormelingen P. 2013. An inordinate fondness? the number, distributions, and origins of diatom species. Journal of Eukaryotic Microbiology 60(4): 414-420. http://doi.org/10.1111/jeu.12047
- Moresco C., Tremarin P. I., Ludwig T. A. V. & Rodrigues L. 2011. Abundant periphytic diatoms in three streams with different anthropic influences in Maringá, Paraná State, Brazil. Brazilian Journal of Botany 34: 359–373. http://doi.org/10.1590/S0100-84042011000300010
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’hara R. B. & Oksanen M. J. 2013. Package ‘Vegan’. Community Ecology Package, Version 2(9): 1–295.
- Pfeiffer F., Gröber C., Blank M., Händler K., Beyer M., Schultze J. L. & Mayer G. 2018. Systematic evaluation of error rates and causes in short samples in next-generation sequencing. Scientific Reports 8: 10950. http://doi.org/10.1038/s41598-018-29325-6
- Pinseel E., Janssens S. B., Verleyen E., Vanormelingen P., Kohler T. J., Biersma E. M., Sabbe K., Van De Vijver B. & Vyverman, W. 2020. Global radiation in a rare biosphere soil diatom. Nature Communications 11(1): 1–12. http://doi.org/10.1038/s41467-020-16181-0
- Pompanon F., Coissac E. & Taberlet P. 2011. Metabarcoding, une nouvelle façon d'analyser la biodiversité. Biofutur 319: 30–32.
- Potapova, M. 2011. New species and combinations in the genus Nupela from the USA. Diatom Research 26: 73–87. http://doi.org/10.1080/0269249X.2011.575111
- Potapova M. 2013. Transfer of Achnanthes decipiens to the genus Nupela. Diatom Research 28: 139–142. http://doi.org/10.1080/0269249X.2012.753114
- Potapova M. & Charles D. F. 2007. Diatom metrics for monitoring eutrophication in rivers of the United States. Ecological Indicators 7: 48–70. http://doi.org/10.1016/j.ecolind.2005.10.001
- Potapova M. G., Ponader K. C., Lowe R. L., Clason T. A. & Bahls L. L. 2003. Small-Celled Nupela species from North America. Diatom Research 18: 293–306. http://doi.org/10.1080/0269249X.2003.9705593
- Reichardt E. 1988. Neue Diatomeen aus Bayerischen und Nordtiroler Alpenseen. Diatom Research 3: 237–244.
- Reimer C. W. 1966. Consideration of fifteen diatom taxa (Bacillariophyta) from the Savannah River, including seven described as new. Notulae Naturae 397: 1–15.
- Rimet F., Abarca N., Bouchez A., Kusber W.H., Jahn R., Kahlert M., Keck F., Kelly M.G., Mann D.G., Piuz A., Trobajo R., Tapolczai K., Vasselon V. & Zimmerman, J. 2018. The potential of high throughput sequencing (HTS) of natural samples as a source of primary taxonomic information for reference libraries of diatom barcodes. Fottea 18: 37–54. http://doi.org/10.5507/Fot.2017.013
- Rimet F., Aylagas E., Borja A., Bouchez A., Canino A., Chauvin C., Chonova T., Jr F. C., Costa F. O., Ferrari B. J. D., Gastineau R., Goulon C., Gugger M., Holzmann M., Jahn R., Kahlert M., Kusber W.-H., Laplace-Treyture C., Leese F., Leliaert F., Mann D. G., Marchand F., Méléder V., Pawlowski J., Rasconi S., Rivera S., Rougerie R., Schweizer M., Trobajo R., Vasselon V., Vivien R., Weigand A., Witkowski A., Zimmermann J. & Ekrem T. 2021a. Metadata standards and practical guidelines for specimen and DNA curation when building barcode reference libraries for aquatic life. Metabarcoding And Metagenomics 5: E58056. Pensoft Publishers. http://doi.org/10.3897/mbmg.5.58056
- Rimet F. & Bouchez A. 2012. Life-forms, cell-sizes and ecological guilds of diatoms in European rivers. Knowledge And Management Of Aquatic Ecosystems 406: 1–14. http://doi.org/10.1051/kmae/2012018
- Rimet F., Chonova T. & Bouchez A. 2021b. Barcoding ADN diatomées: Complétion de la bibliothèque de référence Diat.barcode. report INRAE OFB, UMR Carrtel. Recherche Data Gouv, Version 1: 1–52. http://doi.org/10.57745/3ei00b
- Rimet F., Gusev E., Kahlert M., Kelly M. G., Kulikovskiy M., Maltsev Y., Mann D. G., Pfannkuchen M., Trobajo R., Vasselon V., Zimmermann J. & Bouchez A. 2019. Diat.barcode, an open-access curated barcode library for diatoms. Scientific Reports 9: 1–12. http://doi.org/10.1038/s41598-019-51500-6
- Rimet F., Pinseel E., Bouchez A., Japoshvili B. & Mumladze L. 2023. Diatom endemism and taxonomic turnover: assessment in high-altitude alpine lakes covering a large geographical range. Science of the Total Environment 871: 161970. http://doi.org/10.1016/j.scitotenv.2023.161970
- Rivera S. F., Vasselon V., Bouchez A. & Rimet F. 2020. Diatom metabarcoding applied to large scale monitoring networks: Optimization of bioinformatics strategies using Mothur software. Ecological indicators 109: 105775.
- Rivera S. F., Vasselon V., Ballorain K., Carpentier A., Wetzel C. E., Ector L., Rimet F. 2018a. DNA metabarcoding and microscopic analyses of sea turtles biofilms: complementary to understand turtle behavior. PLoS One 13: E0195770. http://doi.org/10.1371/journal.pone.0195770
- Rivera S. F., Vasselon V., Jacquet S., Bouchez A., Ariztegui D. & Rimet, F. 2018b. Metabarcoding of lake benthic diatoms: from structure assemblages to ecological assessment. Hydrobiologia 807: 37–51. http://doi.org/10.1007/s10750-017-3381-2
- Ruck E. C., Nakov T., Alverson A. J. & Theriot E. C. 2016. Phylogeny, ecology, morphological evolution, and reclassification of the diatom orders Surirellales and Rhopalodiales. Molecular Phylogenetics And Evolution 103: 155–171. http://doi.org/10.1016/j.ympev.2016.07.023
- Rumrich U., Lange–Bertalot, H. & Rumrich M. 2000. Diatoms of the Andes (from Venezuela to Patagonia/Tierra del fuego). Iconographia Diatomologica 9: 1–673. http://doi.org/10.1016/J.Scitotenv.2016.03.173
- Schneck F., Torgan L.C. & Schwarzbold A. 2008. Diatomáceas epilíticas Em riacho de altitude no sul do brasil. Rodriguésia 59: 325–338.
- Silvestro D. & Michalak I. 2012. Raxmlgui: a graphical front-end for RAXML. Organisms Diversity & Evolution 12: 335–337. http://doi.org/10.1007/s13127-011-0056-0
- Skibbe O., Zimmermann J., Kusber W.H., Abarca N., Buczko K. & Jahn R. 2018. Gomphoneis Tegelensis Sp. nov.(Bacillariophyceae): a morphological and molecular investigation based on selected single cells. Diatom Research 33: 251–262. http://doi.org/10.1080/0269249X.2018.1518835
- Stamatakis A., Ludwig T. & Meier H. 2005. Raxml-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics (Oxford, England) 21: 456–463. http://doi.org/10.1093/bioinformatics/bti191
- Takano Y. & Horiguchi T. 2006. Acquiring scanning electron microscopical, light microscopical and multiple gene sequence data from a single dinoflagellate cell 1. Journal of Phycology 42: 251–256. http://doi.org/10.1111/j.1529-8817.2006.00177.x
- Tapolczai K., Vasselon V., Bouchez A., Stenger-Kovács C., Padisák J. & Rimet F. 2019. The impact of OTU sequence similarity threshold on diatom-based bioassessment: a case study of the rivers of Mayotte (France, Indian Ocean). Ecology and Evolution 9: 166–179. http://doi.org/10.1002/ece3.4701
- Tremarin P.I., Freire E.G. Bertolli L.M. & Ludwig T.A.V. 2009. Catálogo das diatomáceas (Ochrophyta– Diatomeae) continentais do estado do paraná. iheringia, Série Botânica 64: 79–107.
- Tremarin P. I., Straube A. & Ludwig T. A. 2015. Nupela (bacillariophyceae) in littoral rivers from south Brazil, and description of six new species of the genus. Fottea 15: 77–93. http://doi.org/10.5507/fot.2015.007
- Trobajo R., Clavero E., Chepurnov V. A., Sabbe K., Mann D. G., Ishihara S. & Cox E. J. 2009. Morphological, genetic and mating diversity within the widespread bioindicator Nitzschia palea (Bacillariophyceae). Phycologia 48: 443–459. http://doi.org/10.2216/08-69.1
- Vasselon V., Domaizon I., Rimet F., Kahlert M. & Bouchez A. 2017a. Application of high throughput sequencing (HTS) metabarcoding to diatom biomonitoring: so DNA extraction methods matter? Freshwater Science 36: 162–177. http://doi.org/10.1086/690649
- Vasselon V., Rimet F., Domaizon I., Monnier O., Reyjol Y. & Bouchez A. 2019. Assessing pollution of aquatic environments with diatoms’ DNA metabarcoding: experience and developments from France water framework directive networks. Metabarcoding and Metagenomics 3: E39646. http://doi.org/10.3897/mbmg.3.39646
- Vasselon V., Rimet F., Tapolczai K. & Bouchez A. 2017b. Assessing ecological status with diatoms DNA metabarcoding: scaling-up on a WFD monitoring network (Mayotte Island, France). Ecological Indicators 82: 1–12. http://doi.org/10.1016/j.ecolind.2017.06.024.
- Verleyen E., Van De Vijver B., Tytgat B., Pinseel E., Hodgson A. D., Kopalova K., Chown L. S., Van Ranst E., Imura S., Kudoh S., Van Nieuwenhuyze W., Antdiat Consortium, Sabbe K. & Vyverman W. 2021. Diatoms define a novel freshwater biogeography of the Antarctic. Ecography 44: 548–560. http://doi.org/10.1111/ecog.05374
- Visco J. A., Apothéloz-Perret-Gentil L., Cordonier A., Esling P., Pillet L. & Pawlowski J. 2015. Environmental monitoring: inferring the diatom index from next-generation sequencing data. Environmental Science & Technology 49: 7597–7605. http://doi.org/10.1021/es506158m
- Wetzel C. E., Potapova M., & Williams D. M. 2022. Synedra phantasma M.H.Hohn (Bacillariophyta, Fragilariaceae) from the Amazon river (South America): its typification and transfer to the genus fragilaria. Notulae Algarum 252: 1–12.
- Youn B. D. & Wang P. 2008. Bayesian reliability-based design optimization using eigenvector dimension reduction (EDR) method. Structural And Multidisciplinary Optimization 36: 107–123. http://doi.org/10.1007/s00158-007-0202-7
- Zimmermann J., Abarca N., Enke N., Skibbe O., Kusber W.H. & Jahn R. 2014. Taxonomic reference libraries for environmental barcoding: a best practice example from diatom research. PLoS One 9: 1–24. http://doi.org/10.1371/Journal.Pone.0108793
- Zimmermann J., Glöckner G., Jahn R., Enke N. & Gemeinholzer B. 2015. Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. Molecular Ecology Resources 15: 526–542. http://doi.org/10.1111/1755-0998.12336
Figure legends for Supplementary data
Supplementary data 1. LM micrographs of Nupela boucheziae sp. nov. Figs 1–23. Valves with raphe and Figs. 1–19. Valves without raphe, from a site called Blanche-Pont de l'Alma in Martinique. Scale bar = 10 µm.
Supplementary data 2. LM micrographs of Epithemia boucheziae sp. nov. Figs 1–10. Valve view. Figs 11–13c. Girdle view, from river Rivière Bras David in Guadeloupe. Scale bar = 10 µm.
Supplementary data 3. SEM micrographs of Epithemia boucheziae sp. nov. Figs 1–2. External valve view. Figs. 3–4 Internal valve view. Scale bars = 10 µm.
Supplementary data 4. Spearman’s correlation between frustule counts of suspected species and percentage of reads of ASVs. (Correlation coefficient: bottom left of the table, p-value: upper right of the table.)
