Abstract
Mycobacterium tuberculosis infection remains one of the major public health issues worldwide. Current qualitative assays (only positive or negative results) do not provide comprehensive information regarding health risk of M. tuberculosis. This study attempted to develop a quantitative assay to measure air concentration of M. tuberculosis in a health care setting.
A total of 22 air samples were taken from the negative pressure isolation rooms of tuberculosis patients. The air was filtered through a Nuclepore filter with sampling time of 8 h. The DNA of M. tuberculosisin these airborne samples was then analyzed by the ABI 7700 real-time quantitative polymerase chain reaction (real-time qPCR) system.
The real-time qPCR method could perform measurements of counts in a dynamic range of over 6 orders with a high sensitivity. The measured M. tuberculosis concentrations varied widely, from 1.43 × 10 copies/m3 to 2.06 × 105 copies/m3. Comparisons among airborne M. tuberculosis levels, sputum smear, results, and sputum culture results showed moderate correlations.
The filter/real-time qPCR method proved extremely sensitive and rapid for quantifying airborne M. tuberculosis. In addition, it is a powerful sampling tool that has potential applications as an investigational device, which might be valuable in conducting studies that validate the efficacy of engineering controls and work practices.
INTRODUCTION
Tuberculosis (TB) remains a major public health problem worldwide due to its high risk of person-to-person transmission, morbidity, and mortality (CitationFalkinham 1996; CitationMartin and Lazarus 2000). Currently, Mycobacterium tuberculosis infects one-third of the world's population (CitationDye et al. 1999), and it is the second-most infectious cause of death worldwide (CitationWHO 2001). Moreover, progressive increases in M. tuberculosis infections are expected, and a worldwide annual incidence of 12 million cases by 2005 is predicted by the World Health Organization (CitationMartin and Lazarus 2000). A sensitive method for monitoring airborne M. tuberculosis is therefore crucial for TB control.
In airborne M. tuberculosis analysis, identification of M. tuberculosis from field sampling by using a culture method is not practical due to slow growth of M. tuberculosis and to contamination by other bacteria and fungi (CitationLoudon et al. 1969; CitationMacher et al. 1992; CitationRiley et al. 1976). Therefore, filter sampling coupled with a commercial DNA detection method based on polymerase chain reaction (PCR) (Amplicor M. tuberculosis PCR test) was proposed to monitor M. tuberculosis in air (CitationSchafer et al. 1998; CitationSchafer and Fernback 1999). This filter-PCR method was also recommended by the National Institute of Occupational Safety and Health for airborne M. tuberculosis measurement (CitationNIOSH 1998). However, this method only qualitatively determines positive or negative responses in a narrow dynamic range (less than four orders of magnitude) (CitationSchafer et al. 1998; CitationSchafer and Fernback 1999). In clinical studies, the most commonly used methods to analyze M. tuberculosis involve using either a smear method for acid-fast bacilli (AFB) staining samples or a culture method. However, these two methods have disadvantages: the AFB staining method has low sensitivity, and the culture method is time consuming.
Rapid identification methods using molecular techniques have been developed recently and utilized in clinical laboratories (CitationBroccolo et al. 2003; CitationCleary et al. 2003; CitationDesjardin et al. 1998; CitationKraus et al. 2001; CitationMiller et al. 2002; CitationShrestha et al. 2003). One such method is the real-time quantitative PCR (real-time qPCR) system, which is a commercially available system designed to decrease the time required for PCR assays by using fluorescent probes to monitor the amplification of the target sequences in real time. One benefit of this real-time qPCR system for clinical assays is that carryover contamination does not occur during postamplification because detection of amplified nucleic acid products is accomplished in a closed system (CitationBroccolo et al. 2003; CitationCleary et al. 2003; CitationDesjardin et al. 1998; CitationKraus et al. 2001; CitationMiller et al. 2002; CitationShretha et al. 2003). Consequently, this molecular method for identification of cultured isolates has been suggested and assessed (CitationRoger van Doorn et al. 2003). However, the use of this technology for airborne M. tuberculosis evaluation needs to be validated.
The aim of this study is to validate this rapid and high-throughput filter/real-time qPCR assay for directly measuring airborne M. tuberculosis in respiratory isolation of TB patients. Moreover, the measured airborne M. tuberculosis levels are also compared with the TB patient status (based on sputum smear and culture results).
MATERIALS AND METHOD
Sampling Location
The respiratory isolation (negative-pressure) rooms of TB patients were investigated. The patients selected for this study were identified by clinical diagnosis of M. tuberculosis pneumonia based on history, symptoms, physical examination, and chest radiographs. These patients were inpatients at the Hospital for Chronic Disease in Taipei and had sputum samples submitted for AFB staining and culture.
Air Sampling
Air samples were taken from the isolation rooms of the selected TB patients. A total of 22 samples from the rooms of six patients were measured from 6 November to 18 December 2003. In this hospital, every patient stayed individually in an isolation room. The age of the six patients ranged from 26 to 74, and five were male and one was female (). For quality control, three samples of field blank were performed in TB patient room. Our results demonstrated that no detectable M. tuberculosis DNA was observed in field blank control (data not shown). In addition, side-by-side duplicate field samples gave comparable results (with relative difference 13%; data not shown).
Table 1 Tuberculosis patients characteristics and airborne M. tuberculosis concentrations
For the filter/real-time qPCR assay, the air in each isolation room was filtered through a 37 mm diameter Nuclepore filter (Costar, Cambridge, MA, USA), which is a track-etched polycarbonate filter consisting of a polycarbonate membrane with straight-through pores of uniform size (0.4 μ m). The filters were supported by cellulose pads and loaded into open-face three-piece plastic cassettes. Before sampling, the filters and support pads were autoclaved and the plastic cassettes were sterilized with ethylene oxide. The sampling flow rate was 22 l/min, and the sampling time was 8 h. The pump and filter apparatus were placed within 1 m from the patient's bed on an adjacent nightstand. The sampling height was 1.2–1.5 m above the floor, so as to be within the human breathzone. After sampling, the Nuclepore filter was removed from the holder and placed in a test tube containing 4 ml of sterile deionized water. The water and filter were then vortexed in the tube for 60 s.
DNA Extraction Method
DNA extraction of the filter elution samples was accomplished using a DNA extraction kit (Hexwater Inc., Germany). First, 200 μl of sample suspension was added to 1 ml of a WB1 lysis buffer, vortexed, and then pelleted by centrifugation at 12,000 rpm (8000 × g) for 1 min. After the supernatant was removed, the pellet was suspended in 200 μ l WB2 lysis buffer, which included the beads at the bottom of the buffer, and then incubated at 55°C for 30 min. The pellet suspension was vortexed for 5 s, spun down, and then boiled at 100°C for 8 min to release the DNA. The samples were again pelleted by centrifugation at 12,000 rpm (8000 × g) for 3 min. Finally, 100 μl supernatant of each sample was saved for PCR and stored at −20°C. All manipulations of the samples were performed in a biological safety cabinet.
ABI 7700 Quantification
Oligonucleotide Primers and Probes
The real-time qPCR assay was done using a forward primer (5′-GGCTGTGGGTAGCAGACC-3′) and reverse primer (5′-CGGGTCCAGATGGCTTGC-3′), which were directed at a 163 bp region of the IS6110 gene sequence (CitationDesJardin et al. 1998). The internal oligonucleotide probe was labeled with fluorescent dyes, namely, 5-carboxyfluorescein (FAM) on the 5′ end and N,N,N′,N′,-tetramethyl-6-carboxyrhodamine (TAMRA) on the 3′ end (5′-[FAM]-TGTCGACCTGGGCAGGGTTCG[TAMRA]-3′). The primer and probe were synthesized using an ABI 7700 (Applied Biosystem Inc., Foster City, CA, USA).
PCR Conditions
In the PCR assay, 50 μl of the PCR mixture solution was placed in each well of a MicroAmp Optical 96-well reaction plate, and then each well was capped with a MicroAmp Optical Cap (PE Biosystems, Foster City, CA, USA). All reagents were from a TaqMan Core PCR Reagent Kit (PE Biosystems, Foster City, CA, USA). The PCR mixture solution consisted of 5 μ l of extracted DNA solution, 25 μl of 2 × TaqMan universal Master Mix, 300 nM forward primer, 300 nM reverse primer, and 250 nM probe. A sequence detector system (ABI Prism 7700; Applied Biosystem, Foster City, CA, USA) was used for amplification and fluorescence measurement. All cycles began with 2 min at 50°C for UNG enzyme incubation and then 10 min at 95°C for AmpliTaq Gold activation. The subsequent PCR assay consisted of 50 cycles, where each cycle involved denaturation at 95°C for 20 s and then annealing and extension at 60°C for 1 min. All samples analyzed using the real-time qPCR were done in triplicate (CV% < 5%).
Standard Curve and Inhibitory Effect
The target DNA standard solution from cloning plasmid was purchased from Mission Biotech (Taipei, Taiwan, R.O.C.). The threshold was set at 10 times the standard deviation of the mean baseline emission calculated for PCR cycles 3–10. The amount of product in a particular reaction mixture solution was determined by interpolation from a standard curve of cycle threshold (Ct) values generated from a known starting concentration of DNA at the same run. Positive (purified dilutions of plasmid DNA) and negative controls were analyzed for each run. The inhibitory effect of the collected air samples on real-time qPCR assay was evaluated by diluting method with 1/10 and 1/100 dilutions of two samples, 2.07× 103 copies/m3 and 1.78 × 105 copies/m3. The results confirmed no inhibitory effect of PCR assay in the collected airborne samples (results not shown).
RESULTS AND DISCUSSION
The aim of this study was to validate the filter/real-time PCR assay for quantitative evaluation of airborne M. tuberculosis in hospital rooms of TB patients. Correlations among airborne M. tuberculosis levels, smear results of sputum samples, and sputum culture results were also assessed.
Dynamic Range and Analytical Sensitivity of Real-Time qPCR Assay
The ABI 7700 TaqMan system has been used to quantity M. tuberculosis DNA in sputum during the treatment of TB patients by using a probe specific for the IS6110 gene region (CitationDesjardin et al. 1998). The IS6110 is a multicopy insertion element found in M. tuberculosiscomplex organisms and has been the target of numerous diagnostic assays. The region targeted for amplification has been shown to be specific for M. tuberculosis (CitationHellyer et al. 1996). Therefore, the IS6110 was also used as a target in our current study.
Figure 1 Calibration curve of known M. tuberculosis DNA concentrations and threshold cycle (Ct) by real-time qPCR assay.
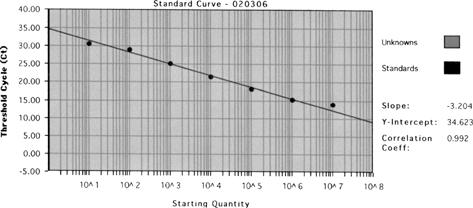
A quantitative assay was developed for IS6110 using the ABI Prism 7700 Sequence Detection System (TaqMan). shows the calibration curve of known M. tuberculosis DNA concentrations and threshold cycle (Ct) by real-time qPCR. The known amounts of M. tuberculosis plasmid DNA yielded Ct values ranging from 20–40 cycles (triplicate standards, with coefficients of variation were less than 1%). The standard curves were linear for over 6 orders of magnitude, ranging from 48 copies/μl to 4.8× 107 copies/μl, with a correlation coefficient (r) value of 0.992. Based on our results, 1 CFU was approximately equal to 253 copies in the real-time qPCR assay by analyzing the fresh cultured M. tuberculosis. The tenfold serial dilutions subjected to the real-time qPCR assay revealed that the sensitivity was less than 48 copies/μl (240 copies/reaction). These results confirm that less than 1 CFU of M. tuberculosis (10 genomes of M. tuberculosis) could be detected by the real-time qPCR assay with high sensitivity, similar to previously reported sensitivity (CitationSchafer et al. 1998; CitationSchafer and Fernback 1999). In conclusion, the filter/real-time qPCR assay developed here could prove useful for rapid identification and quantification of airborne M. tuberculosis.
Airborne M. tuberculosis in TB Patient Rooms
Physical factors of each respiratory isolation room was measured, namely, airflow rate, temperature, relative humidity, and air pressure difference (□). summarizes these measured factors. All six of the patient rooms met the recommended values of air change per hour (ACH; ACH > 8–12) and wind velocities, although two did not meet the recommended pressure difference (> 8 pa).
In our study, the filter/real-time qPCR assay was able to detect and measure the concentration of airborne M. tuberculosis in the TB patient rooms. To our knowledge, this is the first report describing the quantification of airborne M. tuberculosis in field samples. It was found that airborne M. tuberculosis concentration varied widely, from 1.43 × 10 copies/m3 to 2.06× 105 copies/m3 (equal to 1.26× 101 tubercle bacilli/m3 to 8.14 × 103 CFU/m3) (as shown in ). Because infectious dose of M. tuberculosis is as low as 1–5 tubercle bacilli (CitationBalasubramanian et al. 1994), according to our data airborne M. tuberculosis levels in the TB patient rooms studied here might pose a health risk to hospital workers and to families of the patients.
Qualitative filter-PCR assay previously detected M. tuberculosis in the isolation rooms of 6 out of 7 patients studied (CitationMastorides et al. 1999). In addition, 60% of the isolation rooms and the outpatient department area in a TB center contained airborne M. tuberculosis (CitationWan et al. 2004). These findings clearly demonstrate that routine monitoring of airborne M. tuberculosis is crucial in TB centers to ensure the safety of the public and health care personnel. Qualitative assays, however, cannot provide precise information about the health risk of M. tuberculosis.
The filter/real-time qPCR assay can provide deeper insight into hospital epidemiology and infection control, as well as M. tuberculosis transmissibility. This system can be used to determine the background M. tuberculosis levels within isolation rooms, outpatient department areas, microbiology laboratories, and tuberculosis clinics, and to assess the potentially infectious M. tuberculosis dose in room air. Our findings demonstrated that sputum smear results and sputum culture results were moderately correlated with measured airborne M. tuberculosis levels (), and that sputum smear and culture results were moderately correlated (r = 0.57, data not shown). Moreover, 19 of 21 smear samples and 18 of 21 culture samples were positive (). The M. tuberculosis concentrations in the rooms of patients with negative smear and culture results were lower than those with positive smear and culture results (). In general, the airborne M. tuberculosis level in each room was correlated with the clinical TB status of patient.
Figure 2 (a) Correlation between airborne M. tuberculosis levels and sputum smear results. −, No colony growth; +, < 50 CFU/plate; ++, 50–100 CFU/plate; +++, 100–200 CFU/plate. (b). Correlation between airborne M. tuberculosis levels and sputum culture results. +, 1–3 per slide; ++, 1–9 per 10 field; +++, 1–9 per field; ++++.
Based on sputum smear and culture results, patient B seemed more seriously ill than patient D. However, airborne M. tuberculosis levels in the room of patient D were the highest among all samples. From our observation and surveillance, patient D never wore a mask and frequently spit sputum (in the first three sampling dates, 13 November, 20 November, and 4 December). After patient D wore a mask and covered his mouth while coughing and speaking, airborne M. tuberculosis level dramatically decreased (11 December and 18 December). The hygienic practice of this patient might play an important role in preventing the spread of M. tuberculosis.
In conclusion, the filter/real-time qPCR assay is highly sensitive (less than 1 CFU of M. tuberculosis) and fast (< 5 h) for quantifying airborne M. tuberculosis. In addition, this quantitative method has potential applications as an investigational device that may be valuable in conducting studies that validate the efficacy of engineering controls and work practices.
Acknowledgments
The authors thank Dr. Shin-Yu Su for his assistance during this investigation. This workwas supported by grant NSC91-2621-Z-002-025 from the National Science Council, Republic of China. Pei-Shih Chen was supported by a graduate scholarship from the same grant during part of this research effort.
Notes
aPressure difference between isolation room and outside.
bTemperature.
cRelative humidity.
dAir change per hour.
eno colony growth; +, <50 CFU/plate; ++, 50–100 CFU/plate; +++, 100–200 CFU/plate.
fSmear (×400 magnification). +, 1–3 per slide; ++, 1–9 per 10 field; +++, 1–9 per field; ++++, >9 per field.
REFERENCES
- Balasubramanian , V. , Wiegeshaus , E. H. , Taylor , B. T. and Smith , D. W. 1994 . Pathogenesis of Tuberculosis: Pathway to Apical Localization . Tubercle Lung Disease , 75 : 168 – 178 . [CSA]
- Broccolo , F. , Scarpellini , P. , Locatelli , G. , Zinngale , A. , Brambilla , A. M. , Cichero , P. , Sechi , L. A. , Lazzarin , A. , Lusso , P. and Malnati , M. S. 2003 . Rapid Diagnosis of Mycobacterial Infections and Quantification of Mycobacterium tuberculosis Load by Two Real-Time Calibrated PCR Assay . J. Clin. Microbiol. , 41 : 4565 – 4572 . [PUBMED] [INFOTRIEVE] [CROSSREF] [CSA]
- Cleary , T. , Roudel , G. , Casillas , O. and Miller , N. 2003 . Rapid and Specific Detection of Mycobacterium tuberculosis by Using the Smart Instrument and a Specific Fluorogenic Probe . J. Clin. Microbiol. , 41 : 4783 – 4786 . [PUBMED] [INFOTRIEVE] [CROSSREF] [CSA]
- Desjardin , L. E. , Chen , Y. , Perkins , M. D. , Teixeira , L. , Cave , M. D. and Eisenach , K. D. 1998 . Comparison of the ABI 7700 System (TaqMan) and Competitive PCR for Quantification of IS6110 DNA in Sputum During Treatment of Tuberculosis . J. Clin. Microbiol. , 36 : 1964 – 1968 . [PUBMED] [INFOTRIEVE] [CSA]
- Dye , C. , Scheele , S. , Dolin , P. , Pathania , V. and Ravaglione , M. C. 1999 . Global Burden of Tuberculosis. Estimated Incidence, Prevalence and Morality by Country . JAMA , 282 : 677 – 686 . [PUBMED] [INFOTRIEVE] [CROSSREF]
- Falkinham , J. O. 1996 . Epidemiology of Infection by Nontuberculosis Mycobacteria . Clin. Microbiol. , 9 : 177 – 215 . [CSA]
- Hellyer , T. , DesJardin , L. E. , Assaf , M. K. , Bates , J. H. , Cave , M. D. and Eisenach , K. D. 1996 . Specificity of IS6110-based Amplification Assays for Mycobacterium tuberculosisComplex . J. Clin. Microbiol. , 34 : 2843 – 2846 . [PUBMED] [INFOTRIEVE] [CSA]
- Kraus , G. , Cleary , T. , Miller , N. , Seivright , R. , Young , A. K. , Spruill , G. and Hnatyszyn , H. J. 2001 . Rapid and Specific Detection of the Mycobacterium tuberculosisComplex Using Fluorogenic Probes and Real-Time PCR . Mol. Cell. Probes , 15 : 375 – 383 . [PUBMED] [INFOTRIEVE] [CROSSREF] [CSA]
- Loudon , R. G. , Bumgarner , L. R. , Lacy , J. and Coffman , G. K. 1969 . Aerial Transmission of Mycobacteria . Am. Rev. Respir. Dis. , 100 : 165 – 171 . [PUBMED] [INFOTRIEVE]
- Macher , J. M. , Alevantis , L. E. , Chang , Y. L. and Lie , K. S. 1992 . Effect of Ultraviolet Germicidal Lamps on Airborne Microorganisms in an Outpatient Waiting Room . Appl. Occupat. Environ. Hygiene , 7 : 505 – 513 . [CSA]
- Martin , G. and Lazarus , A. 2000 . Epidemiology and Diagnosis of Tuberculosis. Recognition of At-risk Patients is Key to Detection . Postgrad. Med. , 108 : 42 – 44 . 47 – 50 . 53 – 54 . [PUBMED] [INFOTRIEVE]
- Mastorides , S. M. , Oehler , R. L. , Greene , J. N. , Sinnott , J. T. , Kranik , M. and Sandin , R. L. 1999 . The Detection of Airborne tuberculosisUsing Micropore Membrane Air Sampling and Polymerase Chain Reaction . Chest , 115 : 15 – 25 . [CROSSREF]
- Miller , N. , Cleary , T. , Kraus , G. , Young , A. K. and Spruill , G. 2002 . Rapid and Specific Detection of Mycobacterium tuberculosisfrom Acid-fast Bacillus Smear-Positive Respiratory Specimens and BacT/ALERT MP Culture Bottles by Using Fluorogenic Probes and Real-Time PCR . J. Clin. Microbiol. , 40 : 4143 – 4147 . [PUBMED] [INFOTRIEVE] [CROSSREF] [CSA]
- National Institute of Occupational Safety and Health (NIOSH) . 1998 . “ NIOS 0900 Method: Mycobacterium Tuberculosis, Airborne ” . In NIOSH Manual of Analytical Methods , USA : Millie P. Schafer .
- Riley , R. L. , Knight , M. and Middlebrook , G. 1976 . Ultraviolet Susceptibility of BCG and Virulent Tubercle Bacilli . Am. Rev. Respir. Dis. , 13 : 413 – 118 .
- Roger , v an , Doorn , H. , Class , E. C. J. , Tampleton , K. T. , van der Zanden , A. G. M. , te Koppele Vije , A. , de Jong , M. D. , Dankert , J. and Kuigper , E. J. 2003 . Detection of a Point Mutation Associated with High-Level Isoniazid Resistance in Mycobacterium tuberculosisby Using Real-Time PCR Technology with 3′-Minor Groove Binder-DNA Probes . J. Clin. Microbiol. , 41 : 4630 – 4635 . [CROSSREF] [CSA]
- Schafer , M. P. and Fernback , J. E. 1999 . Detection and Characterization of Airborne Mycobacterium tuberculosisH37Ra Particles, a Surrogate for Airborne Pathogen M. tuberculosis . Aerosol Sci. Technol , 30 : 161 – 173 . [CROSSREF]
- Schafer , M. P. , Fernback , J. E. and Jensen , P. A. 1998 . Sampling and Analytical Method Development for Qualitative Assessment of Airborne Mycobacterial Species of Mycobacterium tuberculosisComplex . AIHA J. , 59 : 540 – 546 . [CROSSREF]
- Shrestha , N. K. , Tuohy , M. J. , Hall , G. S. , Reischl , U. , Gordon , S. M. and Procop , G. W. 2003 . Detection and Differentiation of Mycobacterium tuberculosis and Nontuberculosis Mycobacterial Isolatesby Real-Time PCR . J. Clin. Microbiol. , 41 : 5121 – 5126 . [PUBMED] [INFOTRIEVE] [CROSSREF] [CSA]
- Wan , G. H. , Lu , S. H. and Tsai , Y H. 2004 . Polymerase Chain Reaction Used for the Detection of Airborne Mycobacterium Tuberculosis in Health Settings . Am. J. Infect. Control , 32 : 17 – 22 . [PUBMED] [INFOTRIEVE] [CROSSREF]
- World Health Organization (WHO) . 2001 . World Health Report 2001 , Geneva, , Swizerland : World Health Organization .