Abstract
Introduction. The tissue microarray (TMA) technique comprises the potential of significantly reducing time and tissue spent on slicing and performing immunohistochemical (IHC) stainings of paraffin-embedded tumor tissue. Tissue heterogeneity is an argument against using TMAs, which has been dealt with by increasing the size and number of cores punched from each tumor. No consensus exists on the most optimal size, number, and position of TMA cores in the donor paraffin block and no information exist regarding agreement between TMA cores from two different paraffin blocks from the same tumor or between TMA cores and biochemical analyses. Patients and methods. A central and a peripheral 1mm core and a whole section from each of 54 paraffin blocks from 27 breast cancers included in a one-institution cohort, and a single 1mm central TMA core, from each breast tumor from 1000 patients included in the DBCG82 b&c trials, were IHC stained for ER, PgR and HER2. In addition, ER and PgR were measured in the DBCG82 b&c trials by a biochemical analysis. Statistical analyses included Kappa statistics, Kaplan-Meier survival curves, Log-rank tests, and Cox regression hazards analyses. Results and conclusion. IHC stainings for ER, PgR, and HER2 showed a substantial agreement between a single 1mm TMA core and the corresponding whole section, between central and peripheral cores, and between cores from two different paraffin blocks from the same tumor. In addition, a fine agreement was found for ER and PgR between IHC stainings of TMA cores and biochemical analyses. Divergence between IHC and biochemical analyses was predominantly due to the chosen thresholds. IHC staining of one 1mm core from each tumor revealed a significant independent prognostic value of PgR and HER2 on overall survival. In conclusion, IHC stainings for ER, PgR, and HER2 of just a single 1mm TMA core seems to be sufficient, as no significant heterogeneity was noticed.
Immunohistochemical (IHC) stainings have traditionally been applied to whole sections of formalin-fixed paraffin embedded tumor tissues, which is a rather time and tissue consuming technique. In 1998, Kononen and colleagues launched the tissue microarray (TMA) technique comprising the potential of significantly reducing the time spent on slicing and staining the sections, the amount of paraffin embedded tissue spent, and the costs related to reagents and staff Citation[1]. Pros and cons for the TMA technology have been extensively debated. The cons have primarily been related to tissue heterogeneity and a possibly lacking reproducibility of the TMA cores. Whereas in advantage of the TMA technology has been argued that staining of a single TMA slide provides a much larger degree of consistency and standardization than immunostaining of hundreds of slides. In addition, that the scoring criteria are more reproducible, as the entire tumor area is used for interpretation in a TMA core, and thereby avoidance of possible subjective selection of a tumor area as within a whole section. Besides, a superior prognostic value from some stainings of TMA cores as compared with whole sections has been reported Citation[2]. It has been argued that tissue heterogeneity is overcome at best by punching several cores from the same tumor as compared with punching larger cores from the same tumor. Increasing the core diameter from 1 to 2 mm will only increase the proportion of the total tumor examined minimally. Whereas, examining additional punches from different places in the tumor may reveal a more precise distribution of the antigen examined. In breast cancer the reproducibility of staining TMA cores for estrogen receptor (ER), progesterone receptor (PgR) and HER2 receptor as compared with whole sections, has already been validated in several studies Citation[2–6]. No consensus has been reached on the number of cores. It seems, though, that one core reveals sufficient information on ER and HER2 Citation[2–6], but several cores may be necessary to achieve sufficient information on PgR status Citation[2]. No consensus exists whether the cores should be taken from the periphery or the centre of the invasive tumor area. Based on stainings for ER, PgR and HER2, Camp et al. reported that cores may be required from both the tumor edge and tumor centre for adequate representation of the tumors IHC profile Citation[3]. Torhorst et al., however, reported that possible heterogeneity or fixation differences between central and peripheral tumor regions did not affect the prognostic impact of ER and PgR Citation[2]. We have not identified any studies examining the agreement between TMA cores from different paraffin blocks from the same tumor. Good agreement between IHC stainings of whole sections and biochemical analyses, either with dextran-coated charcoal assays (DCC) or solid phase enzyme immunoassays (EIA) has been reported in several studies Citation[7]. We have not identified any studies, though, examining agreement between ER and PgR status when obtained by IHC stainings of TMA cores as compared with biochemical analyses applied to tumor tissue extracts. Several studies have shown good agreement between HER2 positive tumors when detected by IHC stainings as compared with FISH analyses, with the largest discrepancy occurring in tumors scored as 2+ by immunohistochemistry Citation[5], Citation[8–10].
The primary purposes of this study were 1) to examine the reproducibility of IHC stainings for ER, PgR, and HER2 of 1mm TMA cores as compared with whole sections, 2) to examine the agreement between ER and PgR status when identified by IHC stainings of TMA cores as compared with biochemical analyses of crushed tumor tissue, and finally 3) to describe the prognostic value of ER, PgR, and HER2 obtained from IHC stainings of a single 1mm TMA core.
Materials and methods
Patients – one institution cohort comparing TMA cores with whole tumor sections
Twenty-seven patients from a consecutive cohort, previously described in detail Citation[11], were selected. From each patient at least two tumor-containing paraffin blocks were collected.
Patients – DBCG82 b&c TMA study
In the period 1982 to 1990, 3083 high-risk Danish breast cancer patients were enrolled in the DBCG82 b&c studies. High-risk was defined as either tumor-positive axillary lymph nodes and/or tumor size larger than 5 cm and/or invasion of tumor to skin or pectoral fascia. All women had a total mastectomy and a partial axillary dissection. A median of 7 lymph nodes was removed from the axilla. The pre-menopausal women were enrolled in the DBCG82 b protocol and were randomized to either radiotherapy + CMF chemotherapy (8 cycles) or to CMF chemotherapy alone (9 cycles) Citation[12]. The post-menopausal women were enrolled in the DBCG82 c protocol and were randomized either to radiotherapy + tamoxifen (30 mg daily/one year) or to tamoxifen alone Citation[13]. Follow-up of the DBCG82 b&c studies has been described in detail previously Citation[14]. The patients went off-study at the time of recurrence, death or occurrence of a new primary cancer, or at the end of 10 years of follow-up. A long-term follow-up was afterwards performed for patients alive with no relapse the first 10 years after treatment and for patients with an isolated loco-regional recurrence (LRR), another cancer or who died within the first 10 years. No long-term follow-up was performed for patients with distant metastases (DM) or contra-lateral cancer (CBC) as first event, or who died from other cause than cancer within the first 10 years. Long-term survival was obtained from the National Civil Registry for all patients in the study.
A subgroup of 1241 patients, with more than 7 lymph nodes surgically removed out of the 3083 patients, was selected for an extended biological update. New approvals from the ethical committee of Aarhus County and the Danish data protection agency were obtained for the biological study. Median potential follow-up time was 17 years. Endpoints to be considered were distant metastases (DM) and overall survival (OS). DM was defined as any failure outside the ipsilateral mammary region and the regional lymph nodes. In case the patient had contralateral breast cancer and subsequent DM, this was not recorded as DM. Histopathological or cytological confirmation of DM was not performed routinely and often the diagnosis of DM was based on clinical and radiological findings. Histological grade was assessed according to the Bloom and Richardson grade Citation[15].
Methods
Formalin-fixed (neutral buffered) paraffin embedded tumor blocks were collected, sliced, and HE stained from 54 paraffin blocks from 27 primary breast tumors included in the single institution cohort and from 1078 out of the 1241 DBCG82 b&c patients with more than 7 lymph nodes surgically removed. Invasive primary tumor was verified and marked, in all 54 sections from the single institution cohort and in sections from 1 000 out of the 1078 DBCG82 b&c patients with available paraffin blocks. One central and one peripheral 1mm core from the 54 paraffin blocks and one central 1mm core from the 1 000 DBCG82 b&c paraffin blocks were transferred to tissue microarrays (TMAs) using a manual tissue microarray machine from Beecher Instruments. TMAs were constructed including only 30–50 tumor cores in a 24×38mm recipient block, thereby maximizing the number of patients with assessable cores. Four µm thick sections were made from the TMAs and the whole paraffin blocks and the sections were stained IHC for ER, PgR, and HER2 receptor. In general, all sections were stored at 4°C until manual staining, which was performed within a week, however, for HER2 no longer than five days went on from slicing to staining. Deparaffinization, blocking of the endogen peroxidase, and epitope retrieval was performed for all three staining procedures. Sections were stained for ER with the mouse monoclonal antibody estrogen receptor (ER) CONFIRMTM clone 6F11 from Ventana. Two hundred µl of prediluted antibody was applied for 30 minutes. Afterwards the sections were incubated with peroxidase conjugated goat anti-mouse immunoglobulin's (DAKO Envision Mouse, K4001, Dako, Glostrup, Denmark) for 30 minutes at room temperature. Sections were then stained with DAB (Dako, K3468) and counterstained with haematoxylin. PgR staining was performed with the mouse monoclonal antibody PgR 636 from Dako at a dilution of 1:2400. The sections were incubated overnight at a temperature of 4°C and were at day two incubated with Envision Mouse, K4001 (Dako, Glostrup, Denmark) for 45 minutes. Subsequently they were stained with NovaRed (VECTOR SK4800) and counterstained with haematoxylin. HER2 immunostaining was performed with the HercepTestTM kit (Dako, Glostrup, Denmark) according to the manufacturer's manual. One hundred µl of the antibody was added to each section, which was incubated for 30 minutes at room temperature. Afterwards the sections were incubated with a visualization reagent for 30 minutes and were finally stained with DAB and counter stained with haematoxylin. Paraffin sections including breast tumor tissue previously defined as positive and negative for ER, PgR, and HER2 were additionally stained with and without antibody. Stainings were scored semi quantitatively using a light microscope. Stainings of whole sections and TMA cores from the 54 paraffin blocks were scored by two different observers, whereas stainings of the 1 000 DBCG82 b&c TMA cores were scored by one observer. If any doubt occurred during scoring of the 1 000 DBCG82 cores, consensus was reached with another observer. Only staining of invasive tumor cells was scored. ER and PgR were defined as positive if nuclear staining was found in more than 10% of the invasive tumor cells. HER2 was scored positive, according to the HercepTest scoring scheme, if strong complete membrane staining was found in more than 10% of invasive tumor (3 + ). In the DBCG82 b&c study TMA cores containing less than 10 invasive tumor cells were not scored. From these tumors an additional whole section was sliced, stained and scored. Furthermore, when scoring ER and PgR, a remark was made for all TMA cores containing few invasive tumor cells. Later, these ER and PgR stained cores with a remark were reviewed, and all TMA cores containing less than 50 invasive tumor cells had an additional whole section cut, stained, and scored. In addition, a FISH analysis was made for tumors that scored 2+ for HER2 receptor in the DBCG82 b&c study (weak to moderate complete membrane staining in more than 10% of invasive tumor cells). The FISH procedure was performed using the HER2 FISH pharmDx™ Kit (Dako, Glostrup, Denmark). In this kit, the Probe Mix consists of a mixture of Texas Red-labeled DNA cosmid clones covering the HER2 amplicon and fluorescein-labeled peptide nucleic acid (PNA) probes for the chromosome 17 centromeric region. The procedure was carried out according to the manufacturer's manual, in brief as follows. After deparaffinization and rehydration the sections were placed in the pre-treatment solution at 95°C for 10 min followed by pepsin treatment at room temperature for 10 min. Ten µl of probe mix was applied, and the sections were immediately sealed with coverslip and rubber cement. The denaturation of probe and target DNA was performed on a heating block in the temperature interval of 82–92°C for 5 min and incubated overnight in a humidified hybridization chamber at 45°C. After removal of the coverslips the sections were washed in stringent wash buffer at 65°C for 10 min followed by buffer washes and dehydration. Fluorescence mounting media including DAPI was applied and the specimens coverslipped. The sections were stored cold and dark until analyzed no later than a week after the staining procedure. The results were analyzed using a 100-W fluorescence microscope with a Texas Red and fluorescein isothiocyanate (FITC) double filter locating the invasive tumor areas at low magnification and reading the signals at high magnification with ×40 or ×63 oil-immersion objectives. FISH results were recorded in a data report form, listing the number of red signals for the HER2 probe, and the number of green signals for the centromere 17 for each nucleus investigated, in a total of 60 gene signals or a minimum of 20 cells for each tumor. In cases of ductal carcinoma with an in situ component, a HE stained section was used to mark the invasive areas, avoiding in situ components. Signals were counted in nuclei with identifiable boundaries. Optimally, only signals distinctly separated from each other were included, but in cases with high levels of amplification the signals formed clusters and the number had to be estimated. The ratio was calculated as the number of signals for the HER2 gene probe divided by the number of signals for the centromere 17. The cut-off point for amplification was defined as a ratio of 2 or more.
In a subset of the DBCG82 b&c patients, a biochemical steroid receptor analysis had been performed previously. Data on ER was available in 580 of the patients and on PgR in 534 of the patients. The ER and PgR had been measured by a Dextran Coated Charcoal (DCC) assay as recommended by the EORTC Citation[16]. Tumors were considered receptor positive if at least 10 fmol/mg cytosol protein was present. In all cases the receptor content was measured in histological verified malignant tissue. The laboratories involved in the DCC analyses participated in DBCG's quality assurance programme for receptor analyses.
Statistics
Agreements between IHC stainings of TMA cores and whole sections and between IHC stainings of TMA cores and biochemical measurements were analyzed using Kappa statistics. Kappa statistics is explained as the chance-corrected proportional agreement, and possible values range from +1 (perfect agreement) via 0 (no agreement above that expected by chance) to −1 (complete disagreement). A rough interpretation of Kappa values rates +0.4 to +0.6 as moderate, +0.6 to +0.8 as substantial, and +0.8 to +1.0 as almost perfect agreement. Kaplan-Meier survival curves were made as a function of ER, PgR, and HER2 and were tested for differences by a log-rank test. Prognostic value of ER, PgR, and HER2 was analyzed with univariate and multivariate analyses by Cox regression. Hazard ratios (HR)s provided on Kaplan-Meier survival probability plots were HRs of overall mortality. Level of significance was set to 5% and all estimated p-values were two-tailed. Statistical calculations were performed using STATA version 8.2.
Results
One institution cohort – IHC stainings of TMA cores and corresponding whole sections
A high reproducibility between a 1mm TMA core and the corresponding whole section was found for the 27 patients, with Kappa values exceeding 0.60 for all three stainings (). The discordant findings were primarily due to negative TMA biopsies and positive whole sections. In addition, a high agreement was found between one central and one peripheral TMA core from the same paraffin block and between TMA cores from two different paraffin blocks from the same tumor. Agreement seemed higher between central biopsies from two different paraffin blocks as compared with peripheral biopsies from two different paraffin blocks. A substantial agreement between all four different TMA cores from the same tumor was found for all three stainings. In addition, an extraordinarily fine agreement between two whole sections from the same tumor was demonstrated for all the three IHC stainings. Adding one, two or three additional TMA cores to the result found from the first TMA core did not increase the number of ER positive or HER2 positive tumors. For progesterone receptor, though, a slight increase in number of PgR positive tumors was found using extra TMA cores (). A high interobserver agreement was found for all three stainings, both when scoring TMA cores and whole sections, with Kappa values exceeding 0.75 and 0.65, respectively.
Figure 1. Immunohistochemical analyses of ER, PgR, and HER2 on TMAs scored by observer 1 (O1) and observer 2 (O2), respectively. Only tumors that were interpretable on all four cores were included in this analysis. The bars on the left in each group (1) give the frequency of tumor positivity obtained in one TMA core (a central core from paraffin block 1). The next bars give the tumor positivity obtained after combining the data from core 1 and core 2 (2), from core 1, core 2, and core 3 (3), and from core 1, core 2, core 3, and core 4 (4).
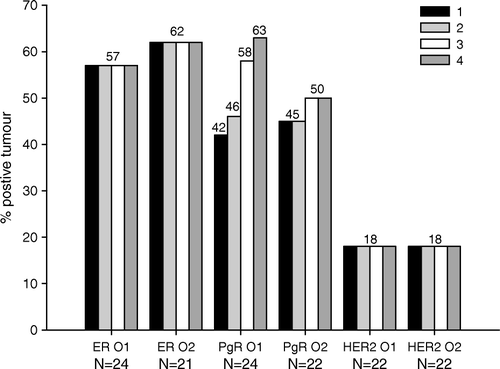
Table I. Kappa values describing comparisons between immunohistochemical stainings for estrogen receptor, progesterone receptor, and HER2 of 108 TMA cores and 54 whole sections from 27 breast carcinomas. Kappa values are presented for observer 1 and observer 2, respectively.
DBCG82 b&c – ER and PgR determined by IHC stainings of TMAs as compared with biochemical analyses
Within the subgroup of 580 patients, with previous biochemical analyses for ER, the frequency of ER positive tumors was 69% when assessed by IHC stainings and 71% when assessed by biochemical analyses (). Overall an agreement of 85% was found for ER status when determined with IHC stainings and biochemical analyses (κ = 0.64). Fifty-six percent (49/88) of the cases discrepant for ER had negative ER status, when assessed by IHC stainings and positive when assessed by biochemical analyses. Within the subgroup of 534 patients with previous biochemical analyses for PgR, 59% of the patients were PgR positive by IHC stainings but 68% by biochemical analyses. The agreement between PgR obtained by IHC stainings and by biochemical analyses was (82%) corresponding to a Kappa value of 0.62. In agreement with the low frequency of PgR positive tumors, when obtained by IHC stainings as compared with biochemical analyses, three-thirds (71/95) of the patients discrepant for PgR had negative PgR status by IHC stainings but positive by biochemical analyses.
Table II. Frequencies of estrogen receptor and progesterone receptor status when determined by immunohistochemical (IHC) stainings and biochemical analyses among high-risk breast cancer patients in the DBCG 82 b&c study.
Further reviewing the discrepant cases, negative by IHC stainings and positive by biochemical analyses, low biochemical values were found for the majority of the patients. Of the 49 ER negative patients by immunohistochemistry but positive by biochemistry, 45 had tumors (92%) with low biochemical values between 10 and 50 fmol/mg. And of the 71 PgR negative tumors by immunohistochemistry but positive by biochemistry 60 (85%) had a biochemical value between 10 and 50 fmol/mg and only 7 (10%) had a high value (>100). Less striking findings were made when reviewing the discrepant cases positive by immunohistochemistry and negative by biochemistry, where 38% (15/39) of the tumors with discordant ER status had a negative biochemical value between 1 and 9 fmol/mg and 25% (9/24) with discordant PgR status had a negative biochemical value between 1 and 9 fmol/mg (data not shown).
Univariate analyses of ER and PgR showed almost equal probability of DM and OS, no matter whether IHC or biochemical analyses were used ().
Table III. Univariate analyses of the prognostic value of ER and PgR for probability of distant metastases (DM) and overall survival (OS), when ER and PgR status is determined by immunohistochemical (IHC) and biochemical (BIO) analyses, respectively.
Similar results were found obtaining information strictly from TMA cores, excluding whole sections).
DBCG82b&c TMA – Prognostic value of ER, PgR, and HER2 after staining a single 1mm central TMA core
In the DBCG82 b&c study, one staining with the respective antibody was performed for each patient's primary tumor, represented by a single 1mm central core. Only 8, 7, and 10 of the cores stained for ER, PgR, and HER2 were completely missing. We achieved information on ER status from 950 and PgR status from 949 of the 1000 patients using a cutpoint of at least 50 invasive tumor cells necessary for scoring the core. After staining whole sections from the remaining 50 and 51 tumors ER status was available in 998 and PgR status in 999 of the patients. Using another cutpoint of at least 10 invasive tumor cells necessary for scoring the core, both stainings were assessable in 976 of the patients. Comparing ER and PgR stainings of each TMA core with the corresponding whole section from the 26 and 27 patients, respectively, with 10–50 invasive tumors cells, agreement on ER status was found in 16/26 (62%) of the patients and on PgR status in 18/27 (67%) of the patients. For 3 and 1 of the patients with diverging ER or PgR status the TMA core was positive and the whole section was negative and for 7 and 8 of the patients the TMA core was negative and the whole section was positive.
Within the total patient group the frequency of ER positive patients was 67% and PgR positive patients was 60%. Kaplan Meier survival probability plots showed superior overall survival for patients with ER positive as compared with ER negative tumors and for patients with PgR positive as compared with PgR negative tumors. The PgR survival difference remained substantial at 15 years of follow-up but the ER survival difference was considerably reduced at 15 years of follow-up (). Negative PgR status was significantly associated with increased overall mortality in a multivariate Cox regression analysis when corrected for randomization, protocol, positive lymph nodes, tumor size, malignancy grade, ER and HER2 (p = 0.003) (). The independent prognostic value of ER status disappeared after correction for PgR.
Figure 2. Kaplan-Meier probability plots of overall survival in high risk breast cancer patients as a function of estrogen receptor, progesterone receptor, and HER2 receptor, respectively.
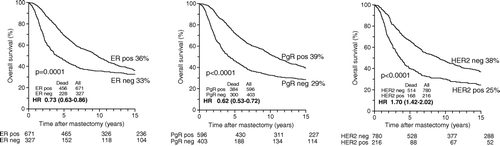
Table IV. Multivariate analysis of overall mortality by Cox regression for 996 high-risk breast cancer patients randomized to +/÷ radiotherapy (RT). Only variables, significantly associated with overall survival in univariate analyses: randomization to +/÷ RT, protocol, positive lymph nodes, tumor size, malignancy grade, ER status, PgR status, and HER2 status, were included in the analysis.
One staining for HER2 receptor of each TMA core included in the DBCG82 b&c study provided us with information on HER2 status for 961 of the 1 000 patients using a cutpoint of at least 10 invasive tumor cells necessary for scoring the slide. After staining whole sections from the remaining 39 patients, HER2 status was available for 996 patients. One hundred and seventy-nine tumors from the DBCG82 b&c study scored 3+, corresponding to a frequency of 18%, and 89 of the tumors scored 2+ corresponding to a frequency of 9%. Among the 89 TMA cores scoring 2+ a successful FISH amplification of a new whole section was performed for 71 (80%) of the tumors. For 37 (52%) of the tumors the ratio of the HER2 probe versus the centromere probe exceeded 2 and they were classified HER2 positive. Thirty-four of the tumors had a ratio of less than 2 and were classified HER2 negative tumors. So, after adding results from FISH analyses, the frequency of HER2 positive tumors increased to 22% (216 patients). A significantly reduced OS was seen for HER2 positive patients as compared with HER2 negative patients with the survival difference remaining substantial after 15 years of follow-up (). Cox multivariate regression analyses showed an independent prognostic value of HER2 on overall mortality when corrected for randomization, protocol, positive lymph nodes, tumor size, malignancy grade, ER and PgR (p = 0.005) ().
Similar results were found when information was obtained strictly from TMA cores and not also obtained from additional whole sections from cores with few invasive tumor cells.
Discussion
We examined tissue heterogeneity related to IHC stainings for ER, PgR, and HER2 and found an almost perfect agreement between whole sections from two different paraffin blocks. A substantial agreement was found between one single 1mm TMA core and the corresponding whole section and between four different TMA cores from the same tumor, as well. In agreement with previous findings we confirmed that differences in stainings of central and peripheral cores from the same paraffin block were not pronounced for ER, PgR, and HER2 Citation[2]. Also corroborating previous findings, we showed that adding results from extra TMA cores did not result in any increase in frequency of ER or HER2 positive tumors for any of the two observers. In PgR positive tumors a slight increase was found after adding extra TMA cores Citation[2], Citation[8]. This was consistent with the fact that PgR discordant cases primarily owed to an underestimation of tumor-positive TMA cores as compared with whole sections in the one institution cohort and as compared with biochemical findings in the DBCG82 b&c study. Apart from a substantial agreement between IHC stainings of TMA cores and biochemical analyses, we found that 75% of the PgR discordant tumors had negative TMA cores and positive biochemical analyses. Notably, the divergence seemed mainly to be due to the chosen thresholds. Applying the most commonly used thresholds both scoring IHC stainings and rating biochemical findings, more than 90% of the ER discordant tumors and 85% of the PgR discordant tumors with negative IHC stainings and positive biochemical analyses had low biochemical receptor content (<50 fmol/mg). It must be emphasized, though, that some divergence is expected, in particular among tumors only weakly positive, as IHC analyses are performed for invasive tumor only but biochemical analyses for a homogenate containing invasive tumor, in-situ tumor components, and normal tissue.
Another concern was that the use of TMA cores might underestimate ER and PgR positivity in tumors with few invasive tumor cells. Comparing TMA cores and whole sections from patients with 10–50 invasive tumor cells in the DBCG82 b&c study we found agreement for 60–70% in both stainings. This was less than found in the one-institution cohort. The discordant tumors had primarily negative TMA cores and positive whole sections and therefore caution should be exercised when scoring TMA cores with few invasive tumor cells.
We chose to analyse reproducibility with Kappa statistics. A limitation of this statistical method is that it only says something about how many cores are misclassified whereas it does not say anything about a possible unequal distribution of the misclassified cores. To handle that limitation we made tables showing a possible unequal distribution.
Torhorst and colleagues have argued that the question to what extent TMA data could reproduce whole section data was much less important than whether clinical-pathological associations could be reproduced or newly detected with TMAs. Hundred percent agreement between TMAs and whole sections was not necessary, due to the large number of patients in the TMA studies Citation[2]. We actually found an identical prognostic value of both ER and PgR applying immunohistochemistry to a single TMA core as compared with biochemical analyses of a homogenized tumor tissue. Moreover, we found a prognostic relevance of all three markers by staining just a single TMA core from each tumor. This is in agreement with Zhang et al. reporting, that one single core per specimen was sufficient to identify associations between biomarkers, including PgR, and clinical-pathological parameters Citation[4]. Another argument for punching more than one core from each patient is to increase the number of interpretable cores. However, in our series of 1 000 patients only 8, 7, and 10 of the cores stained for ER, PgR, and HER2 respectively were missing, because they had been ripped completely off the TMA, the remaining were missing due to few invasive tumor cells. Staining an extra core from the same tumor may predominantly increase the number of interpretable cores with the very small number of cores missing, as they had been ripped off the TMA, and the small number of cores that had been punched from a non-invasive area, by mistake. Consequently, the number of interpretable cores may be increased only minimally after staining an additional core from tumors, with only few distinctly localized invasive tumor cells. In conclusion, staining an extra core from 1 000 patients for ER, PgR, and HER2 may add information on 1% at best 2% extra patients.
A frequency of 18% HER2 positive tumors when assessed by IHC stainings, and 22% after applying FISH analyses to tumors scored as 2+, may seem low, in particular compared with FISH analyses, performed in early studies of primarily high-risk cohorts and patients with metastatic disease, suggesting as many as 30% of breast cancers showed HER2 amplification Citation[17], Citation[18]. However, in two large consecutive cohorts of several thousand patients 19–22% of tumors displayed HER2 amplifications and 11% was scored as 3+ by immunohistochemistry Citation[19–21]. In addition, Pritchard et al. reported for 639 node-positive patients, that 26% had HER2 amplifications and 18–20% were determined as positive with IHC stainings only Citation[22]. It might be argued that it is methodological wrong to supplement our TMA results with whole sections (approximately 5% with few invasive tumor cells) as well as have performed FISH analyses on whole sections. It was a decision that was made in order to increase the number of patients in the study and to optimize the quality of the information obtained from these last patients. All analyses were made, excluding these patients, as well, and as expected comparable results were found. Besides, we decided to use whole sections for FISH analyses in part to increase the success rate of valuable samples, but primarily to minimize the amount of tissue wasted. More than 900 cores would have been wasted, as all the cores staining with 2+ were on different TMAs. Of the 71 tumors scored as 2+ by immunohistochemistry and with successful FISH analyses 52% were amplified using a threshold of the HER2 probe to the centromere probe ratio of at least 2. This is in agreement with findings by Zhang et al. reporting amplification by FISH of 63% of tumors scored as 2+ by immunohistochemistry Citation[5].
Conclusion
No significant heterogeneity was found in this study staining 1mm TMA cores for ER, PgR and HER2, no matter whether the cores were punched centrally or peripherally, were from different paraffin blocks from the same tumor, or were compared with whole sections or results based on biochemical analyses. A tendency towards an underestimation of PgR positive TMA cores was seen, but the prognostic value of ER and PgR was comparable in IHC stainings of TMA cores and in biochemical analyses. Actually a prognostic value was found for all three receptors after stainings of one single core biopsy. This study generally supports the idea that staining of only a single 1mm core biopsy for rather frequently and homogeneously expressed markers, such as ER, PgR, and HER2 receptors, seems to be sufficient to reveal associations between biomarkers and clinical outcome. This is an important issue as tumor size at diagnosis is decreasing whereas the need for prognostication and prediction is increasing.
Acknowledgements
This study was supported by grants from the Danish Cancer Society, the University of Aarhus, the Danish Medical Research Council, and M. L. Jørgensen and Gunnar Hansen's foundation. The authors thank Mogens M. Johannesen, Stine Walther Nielsen, Tine Bovtrup and Birthe Hermansen for excellent technical assistance.
References
- Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998; 4: 844–7
- Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Köchli OR, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol 2001; 159: 2249–56
- Camp RL, Charette LA, Rimm DL. Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000; 80: 1943–9
- Zhang D, Salto-Tellez M, Putti TC, Do E, Koay ES. Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod Pathol 2003; 16: 79–84
- Zhang D, Salto-Tellez M, Do E, Putti TC, Koay ES. Evaluation of HER-2/neu oncogene status in breast tumors on tissue microarrays. Hum Pathol 2003; 34: 362–8
- Henriksen KL, Rasmussen BB, Lykkesfeldt AE, Moller S, Ejlertsen B, Mouridsen HT. Semi-quantitative scoring of potentially predictive markers for endocrine treatment of breast cancer: A comparison between whole sections and tissue microarrays. J Clin Pathol 2007; 60: 397–404
- Andersen J. Determination of estrogen receptors in paraffin-embedded tissue. Techniques and the value in breast cancer treatment. Acta Oncol 1992; 31: 611–27
- Bhargava R, Lal P, Chen B. Feasibility of using tissue microarrays for the assessment of HER-2 gene amplification by fluorescence in situ hybridization in breast carcinoma. Diagn Mol Pathol 2004; 13: 213–6
- Hoang MP, Sahin AA, Ordonez NG, Sneige N. HER-2/neu gene amplification compared with HER-2/neu protein overexpression and interobserver reproducibility in invasive breast carcinoma. Am J Clin Pathol 2000; 113: 852–9
- Clinical laboratory assays for HER-2/neu amplification and overexpression: Quality assurance, standardization, and proficiency testing. Arch Pathol Lab Med 2002;126:803–8.
- Offersen BV, Sorensen FB, Knoop A, Overgaard J. The prognostic relevance of estimates of proliferative activity in early breast cancer. Histopathology 2003; 43: 573–82
- Overgaard M, Hansen PS, Overgaard J, Rose C, Andersson M, Bach F, et al. Postoperative radiotherapy in high-risk premenopausal women with breast cancer who receive adjuvant chemotherapy. Danish Breast Cancer Cooperative Group 82b Trial. N Engl J Med 1997; 337: 949–55
- Overgaard M, Jensen MB, Overgaard J, Hansen PS, Rose C, Andersson M, et al. Postoperative radiotherapy in high-risk postmenopausal breast-cancer patients given adjuvant tamoxifen: Danish Breast Cancer Cooperative Group DBCG 82c randomised trial. Lancet 1999; 353(9165)1641–8
- Nielsen HM, Overgaard M, Grau C, Jensen AR, Overgaard J. Study of failure pattern among high-risk breast cancer patients with or without postmastectomy radiotherapy in addition to adjuvant systemic therapy: Long-term results from the Danish Breast Cancer Cooperative Group DBCG 82 b and c randomized studies. J Clin Oncol 2006; 24: 2268–75
- Bloom HJ, Richardson WW. Histological grading and prognosis in breast cancer; A study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer 1957; 11: 359–77
- Revision of the standards for the assessment of hormone receptors in human breast cancer; report of the second E.O.R.T.C. Workshop, held on 16–17 March, 1979, in the Netherlands Cancer Institute. Eur J Cancer. 1980;16:1513–5.
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235(4785)177–82
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989; 244(4905)707–12
- Owens MA, Horten BC, Da Silva MM. HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer 2004; 5: 63–9
- Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, Ellis GK, et al. HER-2 testing in breast cancer using parallel tissue-based methods. JAMA 2004; 291: 1972–7
- Wolff, AC, Hammond, ME, Schwartz, JN, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. J Clin Oncol 2006.
- Pritchard KI, Shepherd LE, O'Malley FP, Andrulis IL, Tu D, Bramwell VH, et al. HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N Engl J Med 2006; 354: 2103–11