?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background: Efficient and accurate methods are needed to automatically segmenting organs-at-risk (OAR) to accelerate the radiotherapy workflow and decrease the treatment wait time. We developed and evaluated the use of a fused model Dense V-Network for its ability to accurately segment pelvic OAR.
Material and methods: We combined two network models, Dense Net and V-Net, to establish the Dense V-Network algorithm. For the training model, we adopted 100 kV computed tomography (CT) images of patients with cervical cancer, including 80 randomly selected as training sets, by which to adjust parameters of the automatic segmentation model, and the remaining 20 as test sets to evaluate the performance of the convolutional neural network model. Three representative parameters were used to evaluate the segmentation results quantitatively.
Results: Clinical results revealed that Dice similarity coefficient values of the bladder, small intestine, rectum, femoral head and spinal cord were all above 0.87 mm; and Jaccard distance was within 2.3 mm. Except for the small intestine, the Hausdorff distance of other organs was less than 9.0 mm. Comparison of our approaches with those of the Atlas and other studies demonstrated that the Dense V-Network had more accurate and efficient performance and faster speed.
Conclusions: The Dense V-Network algorithm can be used to automatically segment pelvic OARs accurately and efficiently, while shortening patients’ waiting time and accelerating radiotherapy workflow.
Introduction
Accurate delineation of organs at the time of patients’ planning-related CT imaging is a crucial requirement for delivering accurate, effective and timely radiation therapy. However, manual segmentation depends mainly on the experience and ability of the imaging radiologist, and imaging results will vary, sometimes considerably, between different radiologists [Citation1]. Differences in the effects of contouring organs at risk in the pelvic area can be substantial and organ-specific, which may affect the treatment plan, often prolonging patients’ wait time and increasing costs [Citation2,Citation3]. Therefore, an efficient and accurate method is urgently needed to automatically segment OAR in a way that accelerates the radiotherapy workflow and decreases patients’ treatment wait time. At present, atlas-based auto-segmentation (ABAS) is commonly used in routine clinical practice [Citation4–7]. This method relies on the spatial correspondence between the two maps by mapping the space where the image to be segmented is located. However, this method is limited by the patient’s body shape, library size, number of CT image layers, registration accuracy, etc. [Citation8–9]. In some cases, the clinical effects do not meet the clinical requirements necessary to guide treatment [Citation10]. In particular, size and shape, texture gray scale and relative position can vary greatly between different individuals owing to natural variability, disease states, and interference from treatment and organ deformation, which may result in extremely low clinical acceptability of pelvic organ images.
Inspired by the success of deep learning in the field of image understanding and analysis, many researchers have adopted deep learning in the processing of medical images in radiotherapy. At present, ideal results are said to be provided by the Convolutional Neural Network (CNN) for 2D image segmentatdion. Ronnenburger et al. [Citation11] used the U Net trained on transmitted light microscopy images (phase contrast and DIC) and won the 2015 ISBI cell tracking challenge for these categories by a large margin. Segmentation of a 512 × 512 image takes less than one second on a recent GPU. Shakeri et al. [Citation12] used the Fully-Convolutional Neural Network (F-CNN) for segmenting sub-cortical structures of the human brain in magnetic resonance imaging (MRI), with no alignment or registration steps at the time of testing; they further improved segmentation results by interpreting the CNN output as potentials of a Markov Random Field (MRF), in which the topology corresponds to a volumetric grid. Alpha-expansion was used to perform approximate inference through imposing spatial volumetric homogeneity on CNN priors. Men et al. [Citation13] first used a deep deconvolutional neural network (DDNN) for target segmentation of nasopharyngeal cancer in CT images and acquired ideal results in both CTV and GTVnx. Similarly, Thong et al. [Citation14] presented a fully automatic framework for kidney segmentation using convolutional networks (ConvNets) trained using a patch-wise approach to predict the class membership of the central voxel in 2D patches in contrast-enhanced CT scans.
Despite these inspiring results, many challenges remain for 3D image segmentation. First, the anatomical environment of 3D medical images is much more complex than that of 2D images, so more parameters are needed to capture the most representative features. Second, training such as 3D CNN often faces various optimization difficulties, which can include overfitting, gradient disappearance or explosion, along with slow convergence speed. Third, insufficient training data make it even more difficult to capture features and train in depth in 3D CNN [Citation15].
Fusion network models such as ResNet [Citation16], DensNet and others achieve their effects by combining various models, which some authors have reported is able to solve the problems of gradients disappearing when the information passes through many layers, as with limited training samples [Citation16–18]. In order to maximize learning ability, the present study explored the use of a Dense V-Network algorithm based on a merger of Dense Net and V-Net. The purposes of this study were to explore the use of a deep learning model that combined two convolutional neural network frameworks, Dense Net and V-Net, to create a Dense V-Network algorithm and test its performance in segmenting pelvic organs at risk.
Material and methods
Model structure
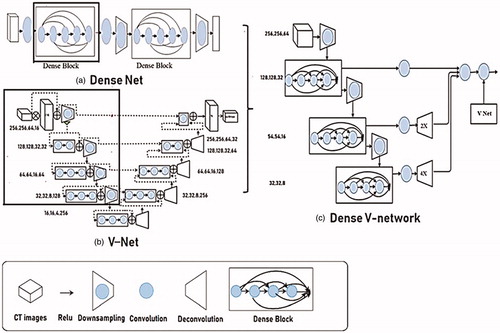
Convolutional neural networks (CNN) use convolutional layers to extract image features, and extract more abstract feature values by deepening network layers. Because convolutional networks can be deeper, more accurate, and efficient to train when they contain shorter connections between layers close to the input and those close to the output, we used the Dense Block structure, as described previously [Citation17]. In Dense Block, each layer gets additional input from all previous layers and passes its own feature map to all subsequent layers in a feed-forward fashion. Whereas conventional convolutional networks with L layers have L connections—one between each layer and its subsequent layer—our network has L(L2 + 1) direct connections. For each layer, the feature-maps of all preceding layers are used as input, and its own feature-maps are used as input in all subsequent layers. EquationEquation (1)(1)
(1) (shown below) shows that this densely connected structure solves the under-fitting phenomenon that occurs during training, and also greatly reduces the number of parameters needed to be trained due to reusing the feature map. The
layer receives the feature-maps of all preceding layers, x0;: : : ; xl−1, as input:
(1)
(1)
where [x0; x1;: : : ; xl−1] refers to the concatenation of the feature-maps produced in layers 0, …, l − 1.
The V-Net network combines 3D convolution and depth residual network, as previously described [Citation19]. As shown in , the perception of the three-dimensional data is effectively improved, and the residual network is used to solve the degradation phenomenon that increases the depth of the training error and accelerates the convergence. The novel objective function is based on Dice coefficient, which is a quantity ranging between 0 and 1, which we aimed to maximize. The dice coefficient D between two binary volumes can be written as
(2)
(2)
where the sums run over the N voxels, of the predicted binary segmentation volume pi
P and the ground truth binary volume gi
G. This formulation of Dice can be differentiated yielding the gradient
(3)
(3)
computed with respect to the j-th voxel of the prediction. Using this formulation, we did not need to assign weights to samples of different classes to establish the right balance between foreground and background voxels, and we obtained much better results than the ones computed through the same network trained by optimizing a multinomial logistic loss with sample re-weighting.
The fused image segmentation model Dense V-Network network absorbs the advantages of Dense Net and V-Net, as shown in . The left part of the Dense V-Network network is a compressed path, which is pooled with a 2*2*2 convolution kernel. Each stage contains one to four convolutional layers, which ensures the flow of information and reduces training parameters through a closed connection. The features extracted from the early stages of the left are passed to the right part, which collects tiny details lost in the compression path and improves the quality of the final contour prediction.
Training data
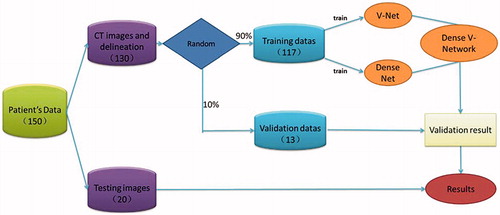
This study enrolled 190 patients with similar pelvic tumors who were admitted to the PLA General Hospital from June 2016 to September 2018. All patients were diagnosed with cervical cancer, including 123 cases that were enrolled after undergoing surgery, with clinical stage determined as IB1-IVA. QAR delineation on the CT images was completed by the attending physician, and then revised and approved by the senior chief physician. In the training of the CNN model, 160 cases were randomly selected as the training set to adjust the parameters of the automatic segmentation model, and the remaining 30 cases were used as test sets to evaluate the performance of the convolutional neural network model.
The images need to be pre-processed and the image size of the model was 320*320*64 or 320*320*32, which is mainly determined according to the distribution area of the organ to be outlined. Dozens of samples can be extracted from each case in this way, so the sample can be expanded several times. During training, the system randomly selects the input area. To prevent a large number of unrelated layers x from participating in the training, the minimum number of OAR involved in the training area will be set. The input image is slightly deformed and rotated by ±10 degrees on the x, y, and z axes. Such preprocessing increases training data, improves the generalization ability of the model, and reduces model parameters. Based on the Atlas segmentation method, this study used automated segmentation software MIM (MIM Software, Inc., Cleveland, OH, USA), and randomly used 80 patients to build a library of maps, and 20 patients to verify.
Model training
To minimize the training period, V-Net and Dense Net networks were trained separately and then frozen when they reached optimal training. The fusion layer was fine-tuned to achieve the best fusion effect. The training process is shown in . The CNN algorithm proposed is based on implementation of the Keras deep learning library, and all budgets are on NVDIA graphics cards to accelerate network training. Model training, evaluation, and error analysis were performed using Tensorflow (Google Brain, Mountain View, CA, USA) and then compiled using the cuDNN compute kernel. The initial learning efficiency was set to 0.0001, the learning rate attenuation factor was 0.0005, the attenuation step size was 1000, and the number of iterations was set to 50,000 for model training.
Evaluation indicators
The accuracy of automatic segmentation was evaluated quantitatively using three parameters: Dice Similarity Coefficient (DSC), Hausdorff Distance (HD) and Jaccard distance (JD).
(1) The Dice similarity coefficient is the degree of similarity between the automatic segmentation result and the physician’s manual segmentation result. A and B are used to represent the point set contained in the two regions. The calculation formula of the DSC is as shown in [Citation4]:
(4)
(4)
where |A∩B| represents the intersection of A and B. The value range of the DSC is 0–1, and the higher the value, the better the segmentation effect.
(2) The Hausdorff distance reflects the largest difference between the two contour point sets. The calculation formula is as shown in [Citation5]:
(5)
(5)
where h(A, B) represents the maximum distance between the point in set A and the point in set B.
The smaller the HD value, the higher the segmentation accuracy.
(3) The Jaccard distance is used to measure the difference between the two sets. The larger the Jaccard distance value is, the lower the sample similarity is. The formula is as shown in formula [Citation6]:
(6)
(6)
Results
Segmentation accuracy and quantitative evaluation of dense V-network algorithm
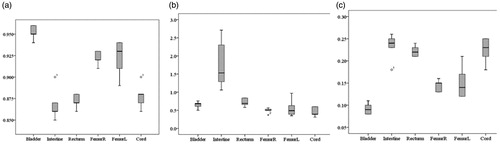
Results of model segmentation algorithm, including the DSC value, Hausdorff distance value and Jaccard distance box pattern, are shown in . The segmentation result is ideal, but the Hausdorff distance of the small intestine is somewhat larger.
To more clearly see the effects of automatic segmentation, the average of the three quantitative parameters of each endangered organ is shown in . The DSC values of the bladder, small intestine, rectum, femoral head and spinal cord are all above 0.85 (average value 0.9) and the Jaccard distance is within 2.3 (average 0.18). With the exception of the intestine, the Hausdorff distance values are all within 9.0 mm (average value 6.1 mm).
Table 1. Segmentation results of pelvic organs at risk (Mean).
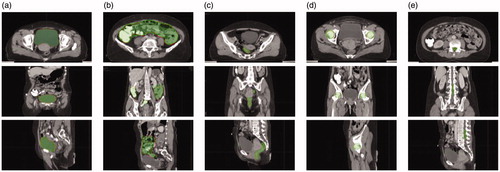
shows the segmentation effects of CT images of cervical cancer, in which the red line indicates the outline of the threatened organ, and the blue area is the result of automatic segmentation by the Dense V-network algorithm. The automatic segmentation result of this model is in good agreement with the manual delineation results.
Evaluation of segmentation effects of each model
After training and testing, the three structures of V-Net, Dense Net and Dense V-Network and comparing them with the MIM software automatic segmentation results, the segmentation effects of the fusion model were evaluated. Evaluation is based on the Dice coefficient value, which is most commonly used to evaluate the accuracy of automatic and semi-automatic segmentation methods. The results are shown in Supplementary Table S1. DSC > 0.7 means that the automatic segmentation area and the manual segmentation area have a high degree of repetition and the segmentation effect is satisfactory, as previously described [Citation20]. Supplementary Table S1 reveals that the automatic segmentation results of the three models are acceptable, and the Dice value of all organs of the fusion model is better than that of the single model.
Discussion
In the present study, a Dense V-Network model was proposed for automatic segmentation of pelvic organs and satisfactory results were realized. Accuracy of automatic segmentation was evaluated based on (1) the degree of deviation of contours by deviation of centroid, minimum average distance and HD, and (2) the differences in volume were evaluated by deviation of volume, sensitivity index, inclusiveness index and JD. Results of the present study have shown that, except for the poor Hausdorff distance of the intestines, the evaluation indexes of the other organs(bladder, rectum, femoral head and spinal cord)were excellent. Also, the delineation effects achieved were significantly better than those of the commonly used Atlas method. Nevertheless, the method of model fusion described in this study requires more time for training and testing than the original models, which is equivalent to the time exchange effect. The total time to perform this method is within three minutes of the original, including segmentation of all organs and post-processing of the results given to the model using an image processing-based method.
Dense V-Networks have been proposed and used for organ segmentation in published studies; however, they are applied only to organs with a simple structure and with a large difference in CT density from neighboring organs such as lung lobes and abdominal organs. However, a better segmentation method has not been established for the pelvic organs owing to their complexity. In particular, the size, morphology, gray scale and relative position of the intestinal tract are affected by individual differences, disease states and previous treatment, making it difficult to create an efficient, effective segmentation method. The Dense V-Network algorithm we developed is based on multi-model fusion for automatic segmentation of endangered organs in radiotherapy, combining the advantages of Dense Net and V-Net network models. Although we developed the Dense V-Network algorithm independently, other authors have introduced a similar model for abdominal organ segmentation [Citation18]. The fused model not only reduces the number of parameters that need to be adjusted in the network and shortens the training time, but also solves the problem caused by the increase of network depth, which improved accuracy and enhanced the training. Overall, the Dense V-Network model quickly and accurately delineated pelvic organs at risk when the training sample was small. However, the results we obtained for the small intestine were poor, likely due to the following: 1. The CT boundary is unclear, making it difficult for the marking physician to sketch; 2. Several physicians drew the intestine and colon together; 3. Some data did not cover the entire intestine, only the part that is closest to the target area, which needed to be dose-evaluated; 4. The shape of the intestine changes due to the degree of organ filling. 5. The small intestine is relatively large so that it magnifies the deviation. These are common problems associated with other auto-segmentation results of soft tissue organs, and solutions must be explored in subsequent research.
Our results for the segmentation effects of all four models (as shown in Supplementary TableS1) shows that the Dense V-Network algorithm has certain advantages compared with the commonly used Atlas method, especially for structures of the bladder and rectum. Quantitative evaluation by DSC value shows that the fused model is superior to single models, the Atlas segmentation method and many international reports. However, if the model is applied to the clinical work of radiotherapy, supervision by physicians will be needed and results will vary accordingly. Then, as the algorithm structure is optimized and the training data increases, the expected results will be improved further.
Several international reports on pelvic endangered organ segmentation have been based on deep learning. Kazemifar et al. [Citation21] used 2D U-Net to directly train the 2D CT gradation transformation to the corresponding feature map of the endangered organ segmentation image, and Dice values of the bladder and rectum were 0.95 and 0.92, respectively, which are better than our results. However, U-Net based on 2D image segmentation does not conform to the 3D characteristics of medical images, and cannot guarantee the information exchange between the upper and lower layers of CT images, which may cause certain deviations in practical applications. Balagopal et al. [Citation22] used the 3D U-Net network to segment the bladder, rectum, and left and right femoral heads in pelvic CT images with Dice values of 0.95, 0.84, 0.96, and 0.95, respectively. The segmentation result of the bladder is comparable to results in the present study, in which the Dice value of the rectum was 0.87, only slightly better than that of Balagopal et al. [Citation22]. Men et al. [Citation23] used the deep dilated convolutional neural network (DDCNN) to automatically segment the bladder, small intestine, rectum, and left and right femoral heads; with Dice values of 0.934, 0.653, 0.62, 0.921, and 0.923, respectively. Results of the present study, especially for the intestine and rectum, are superior to those achieved by DDCNN. In the present study, the fusion model Dense V-network algorithm automatically segmented the pelvic organs-at-risk. Except for the Hausdorff distance of the intestine, our results exceeded those of most other reports.
Convolutional neural networks based on multi-layer supervised learning are said to have good fault tolerance, adaptability and weight sharing [Citation18,Citation21–23]. The trained model is simple to operate, provides reliable results, and is suitable for clinical applications. However, convolutional networks have certain limitations that may jeopardize organ segmentation, and meeting clinical requirements can be difficult. Therefore, we attempted to integrate different networks to improve the effects and accuracy. Overall, CNN has achieved satisfactory results in the fields of two-dimensional image segmentation and image recognition. Convolutional networks are reported to be substantially deeper, more accurate, and efficient to train if they contain shorter connections between layers close to the input and those close to the output. Therefore, we used the Dense Block, as previously described [Citation17], in which each layer gets additional input from previous layers and passes its own feature map to all subsequent layers in a feed-forward fashion. The Dense Net network is usually used for segmentation of OARs in radiation therapy. Gibson et al. [Citation18] used dilated convolutional networks with dense skip connections to segment the liver, pancreas, stomach and esophagus from abdominal CT without image registration, showing the potential to support image-guided navigation in gastrointestinal endoscopy procedures. Nasr-esfahani et al. [Citation24] also used Dense Net in lesion segmentation in non-dermoscopic images and acquired a perfect Dice score, outperforming state-of-the-art algorithms in segmentation of skin lesions based on the Dermquest dataset. Other investigators have adopted dense connectivity to help the learning process by including deep supervision [Citation25]. Highly competitive results were obtained when evaluating their approach compared to data from the MICCAI Grand Challenge on 6-month infant Brain MRI Segmentation (iSEG).
V-Net network can be regarded as a three-dimensional U-Net network, which is widely used in 3D medical image segmentation. Casamitjana et al. [Citation26] segmented brain tumors with a cascade of two CNNs inspired by the V-Net architecture, reformulating residual connections and making use of ROI masks to constrain the networks to train only on relevant voxels. In addition, V-Net is used to segment prostate [Citation19], infant brain [Citation27], white matter hyperintensities [Citation28], and so on. Different models have different effects on endangerment of organs with different structures and we cannot expect a particular model to delineate all organs better than other models. Drawing on the idea of using multiple weak classifiers to realize a strong classifier, we combined well-performing single models to construct an even better, more effective combination model. The merged Dense V-Network model realizes the complementary advantages of the two included network structures, which reduces the need to adjust parameters, reduces redundancy, saves memory space occupied by training, and reduces over-fitting. The combination of the two effective structures solves the problem of gradient disappearance or explosion when training three-dimensional data, improves the convergence time, and simply increases the network depth to improve network performance without increasing the training set.
Limitations
Several limitations must be noted. First, our efforts to validate the model may have been influenced by the participation of all authors, without blinding, in the manual segmentations while the algorithm was being developed and tested. Also, we did not optimize the algorithm parameters for Dense Net and V-Net separately from development of the combined Dense V-Net, and performance data may have been underestimated for the two component methods. We could not evaluate the clinical utility of the method directly, only in the research setting, especially the use of the algorithm for segmentation in image-guided endoscopic navigation in the pelvic region. Additional prospective studies are needed to confirm our initial findings regarding efficiency and accuracy of the method, and to directly compare performance with other methods so that performance can be optimized.
Conclusions
In conclusion, the Dense-V-Network algorithm can be used to automatically segment pelvic OARs accurately and efficiently, while shortening patients’ wait time and accelerating radiotherapy workflow. Overall, the optimized algorithm structure and increased training data are able to improve the clinical results of segmentation.
Supplemental Material
Download MS Word (12.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Duane FK, Langan B, Gillham C, et al. Impact of delineation uncertainties on dose to organs at risk in CT-guided intracavitary brachytherapy. Brachytherapy. 2014;13:210–218.
- Sykes J. Reflections on the current status of commercial automated segmentation systems in clinical practice. J Med Radiat Sci. 2014;61:131–134.
- Nelms BE, Tomé WA, Robinson G, et al. Variations in the contouring of organs at risk: test case from a patient with oropharyngeal cancer. Int J Radiat Oncol Biol Phys. 2012;82:368–378.
- Greenham S, Dean J, Fu CK, et al. Evaluation of atlas-based auto-segmentation software in prostate cancer patients. J Med Radiat Sci. 2014;61:151–158.
- Anders L, Stieler F, Siebenlist K, et al. A validating study of ABAS: an atlas-based auto-segmentation program for delineation of target volumes in breast and anorectal cancer. Intl J RadiatOncolBiol Phys. 2010;78:S836.
- You HQ, Dai Y, Bai PG, et al. The application of atlas-based auto-segmentation (ABAS) software for adaptive radiation therapy in nasopharyngeal carcinoma. J Mod Oncol. 2017;20:3334–3346.
- Xu Z, Burke RP, Lee CP, et al. Efficient multi-atlas abdominal segmentation on clinically acquired CT with SIMPLE context learning. Med Image Anal. 2015;24:18–27.
- Gresswell S, Renz P, Werts D, et al. (P059) Impact of increasing atlas size on accuracy of an atlas-based auto-segmentation program (ABAS) for organs-at-risk (OARS) in head and neck (H&N) cancer patients. Int J Radiat Oncol Biol Phys. 2017;98:E31.
- Sjöberg C, Lundmark M, Granberg C, et al. Clinical evaluation of multi-atlas-based segmentation of lymph node regions in head and neck and prostate cancer patients. RadiatOncol. 2013; 8:229.
- Xu Z, Lee CP, Heinrich MP, et al. Evaluation of six registration methods for the human abdomen on clinically acquired CT. IEEE Trans Biomed Eng. 2016;63:1563–1572.
- Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation MICCAI 2015, part III. In: Navab N, Hornegger J, Wells W, et al., editors. Medical image computing and computer-assisted intervention – MICCAI 2015. Cham: Springer International Publishing; 2015. p. 234–241.
- Shakeri M, Tsogkas S, Ferrante E, et al. Sub-cortical brain structure segmentation using F-CNNs. Proceedings of the IEEE 13th International Symposium on Biomedical Imaging (ISBI); 2016 Apr 13–16; Prague, Czech Republic. 2016. p. 269–272.
- Men K, Chen X, Zhang Y, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol. 2017; :315.
- Thong W, Kadoury S, Piche N, et al. Convolutional networks for kidney segmentation in contrast-enhanced CT scans. Comput Methods Biomech Biomed Eng Imaging Vis. 2018;6:277–282.
- Dou Q, Yu L, Chen H, et al. 3D deeply supervised network for automated segmentation of volumetric medical images. Med Image Anal. 2017;41:40–54.
- He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. CVPR. 2015:770–778.
- Huang G, Liu Z, Van der Matten L, et al. Densely connected convolutional networks. CVPR. 2016:2261–2269.
- Gibson E, Giganti F, Hu Y, et al. Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans Med Imaging. 2018;37:1822–1834.
- Milletari F, Navab N, Ahmadi SA. V-Net: fully convolutional neural networks for volumetric medical image segmentation. Proceedings of the IEEE International Conference on 3D Vision; 2016 Oct 25–28; Stanford, CA. 2016. p. 565–571.
- Andrews S, Hamarneh G. Multi-region probabilistic dice similarity coefficient using the aitchison distance and bipartite graph matching. arXiv. 2015.
- Kazemifar S, Balagopal A, Nguyen D, et al. Segmentation of the prostate and organs at risk in male pelvic CT images using deep learning. Biomed Phys Eng Express. 2018;4.
- Balagopal A, Kazemifar S, Nguyen D, et al. Fully automated organ segmentation in male pelvic CT images. Phys Med Biol. 2018;63:245015.
- Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44:6377–6389.
- Nasr-Esfahani E, Rafiei S, Jafari MH, et al. Dense pooling layers in fully convolutional network for skin lesion segmentation. Comput Med Imaging Graph. 2019;78:101658.
- Dolz J, Ayed IB, Yuan J, et al. Isointense infant brain segmentation with a hyper-dense connected convolution. Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI); 2018 Apr 4–7; Washington, DC. 2018.
- Casamitjana A, Catà M, Sánchez I, et al. Cascaded V-net using ROI masks for brain tumor segmentation. International MICCAI Brain Lesion Workshop. Barcelona (Spain): Springer International Publishing; 2017. p. 381–391.
- Casamitjana A, Sanchez I, Combalia M, et al. Augmented V-Net for infant brain segmentation. Miccai Grand Challenge on 6-Month Infant Brain MRI Segmentation, MICCAI Brain Lesion Workshop. Barcelona (Spain): Springer International Publishing; 2017.
- Casamitjana A, Sanchez I, Combalia M, et al. Augmented V-Net for white matter hyperintensities segmentation. WMH Segmentation Challenge, MICCAI Brain-Lesion Workshop. Barcelona (Spain): Springer International Publishing; 2017.