Abstract
In North Island New Zealand three species of tree wētā (Hemideina) have narrow regions of overlap. Using detailed measurements of chromosomes we compared the karyotypes of Hemideina thoracica with those of Hemideina crassidens and Hemideina trewicki. Although H. thoracica and H. trewicki have the same diploid number (2n = 17 [XO], 18 [XX]), distinct from H. crassidens (2n = 15 [XO] 16 [XX]); the karyotypes of H. trewicki and H. crassidens are more similar to each other than either is to H. thoracica. Elements within each karyotype were identified that are species specific and will aid identification of putative hybrids. Quantitative cytogenetics was used to identify the sex chromosome for H. crassidens and H. trewicki, which in contrast to previous inferences, is most likely the fifth-longest metacentric chromosome in H. crassidens, and the third-longest metacentric chromosome in H. trewicki.
Introduction
Chromosomes are the fundamental units of inheritance. Although most recent population genetic studies focus on DNA sequence variation within specific genes, or at non-coding loci, chromosomes remain a valuable source of information, particularly when looking at hybridisation and introgression between populations (Searle Citation1993; Morgan-Richards et al. Citation2009 and references therein). Chromosome rearrangements have been implicated in hybrid disadvantage, and via linkage involved in speciation (Sites & Moritz Citation1987; Rieseberg Citation2001; Kirkpatrick & Barton Citation2006; Kirkpatrick Citation2010). In sexually reproducing organisms, successful gamete formation is mediated by chromosomal homology, even though nucleotide sequences of individual gene loci may differ. Karyotypes can impose constraints on genetic exchange where homology is not maintained. Abnormal karyotypes can result from de novo mutations or hybridisation (Kirkpatrick & Barton Citation2006; Kirkpatrick Citation2010), and can result in breakdown in gene flow via reduced fertility, or linkage of alleles that might facilitate speciation (Rieseberg Citation2001). Quantitative cytogenetics allows comparison of karyotypes and provides the ability to identify chromosomes that are unique to species and potentially important markers of gene flow (Sessions Citation1996).
Tree wētā (Insecta: Orthoptera: Anostostomatidae: Hemideina) are endemic to New Zealand and are an important component of native New Zealand forest ecosystems (Griffin et al. Citation2011). Chromosomally, Hemideina has attracted interest because two of the seven species are known to comprise more than one chromosome race (Morgan-Richards Citation1997, Citation2000). These races occupy discrete geographic ranges () but there is evidence of intraspecific introgression where some of them meet (Morgan-Richards Citation1997; Morgan-Richards et al. Citation2001a,Citationb, Citation2003). In contrast, not all of the described species of Hemideina have karyotypes that can be distinguished using light microscopy (Morgan-Richards & Gibbs Citation2001). The sex chromosome (X) in all Hemideina species has been identified as one of the large metacentric chromosomes (Morgan-Richards Citation1997) that always occur in odd numbers in males, but are paired in females. However, as all tree wētā karyotypes have more than three large metacentric chromosomes of similar size, the exact identity of the sex chromosome is uncertain.
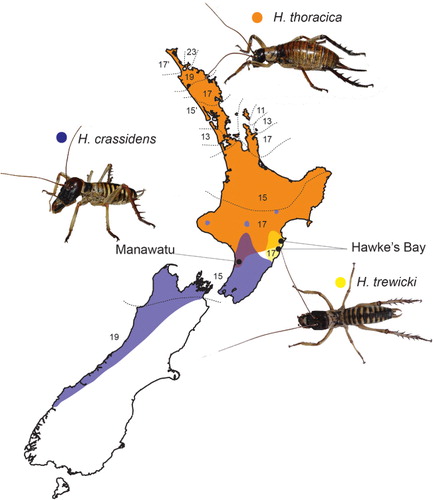
Hemideina crassidens (Blanchard) () has two known chromosome races, one with a diploid (2n) number of 15 (XO males)/16 (XX females) and one race with 19/20 chromosomes. The difference in diploid number between the sexes is due to an XO sex determination system; but the variation in chromosome number that differentiates the two races is the result of the fusion/fission of two autosomes at their centromere resulting in different diploid numbers and distinct karyotypes (Morgan-Richards Citation2000). The two chromosome races of H. crassidens are not distinguished by morphological or allozyme characters (Morgan-Richards et al. Citation1995) and can successfully produce F1 offspring in laboratory crosses (Morgan-Richards Citation2000). Hemideina thoracica (White) has nine known chromosome races, which have karyotypes with between 12 and 24 chromosomes (female 2n). The chromosome races of H. thoracica interbreed in narrow regions of contact and limited introgression of mitochondrial and nuclear alleles has been shown (Morgan-Richards Citation1997; Morgan-Richards et al. Citation2000, Citation2003; Morgan-Richards et al. Citation2001b). This suggests that chromosomal differences alone are not sufficient to cause reproductive isolation between chromosome races in this genus.
The seven species of Hemideina have broadly parapatric ranges, with narrow regions of overlap. Hybridisation is possible between some Hemideina species (Morgan-Richards et al. Citation2001a) although all evidence is of F1 hybrids only based on morphology of wild specimens or captive observations. As yet, it is unknown if viable F2 hybrids are produced leading to introgression of alleles between Hemideina species. In one species pair (Hemideina ricta and Hemideina femorata) the production of F1 hybrids has not led to genetic introgression (Morgan-Richards & Townsend Citation1995).
The present study examines the three parapatric species of Hemideina in the North Island of New Zealand, all of which come into contact at the borders of their range. The 17/18 chromosome race of H. thoracica occurs in the southern part of the North Island of New Zealand where it overlaps with the 15/16 chromosome race of H. crassidens in the Manawatu region (). The 17/18 chromosome race of H. thoracica also occurs in Hawke's Bay where it overlaps with Hemideina trewicki (Morgan-Richards), which has only one known chromosome race (2n = 17/18, ). Putative F1 hybrids have been identified based on body colouration and karyotype in Hawke's Bay (Morgan-Richards et al. Citation1995), but quantitative analyses of chromosomes are required to confirm these identifications, and to investigate whether F1 hybrids are fertile.
We document karyotype variation for the three wētā species at the regions where H. thoracica and H. crassidens overlap in the Manawatu, and H. thoracica and H. trewicki overlap in Hawke's Bay. To increase karyotype resolution the analysis uses chromosome measurements to describe both relative chromosome length, and centromeric position based on arm-length ratios. This quantitative approach to karyotype characterisation is important for studying chromosomal variation within species and comparing among them. This tool will help identification of chromosomes that are unique to each species and so provide a robust method for detection of F1 hybrids and possible backcross individuals.
Methods
Ten specimens each of H. thoracica and H. crassidens were collected from Manawatu, where they are sympatric (Kahuterawa Valley GPS: 40.47184S, 175.60943E; ). Nine H. thoracica and thirteen H. trewicki were collected from Hawkes Bay (Mohi Bush Scenic Reserve: 39.85S, 176.88333E and Elsthorpe Scenic Reserve: 39.91881S, 176.8042E; ). Only males were used for this study as chromosome numbers differ between sexes due to their XO sex determination system, and males provide tissue with high mitotic rates. Species were identified using their unique colour combinations of pronotum and abdomen (Ramsay & Bigelow Citation1978). For each wētā at least three mitotic cells were identified and photographed, and then the chromosomes were counted to confirm karyotype. One mitotic chromosome spread from each wētā was then measured for analysis. Two wētā from a single population (Hawkes Bay H. thoracica) were studied for intraspecific variation by photographing and analysing 10 spreads for each.
After euthanasia with ether, testes were removed and placed in hypotonic solution (1:4 insect saline:water) for 15 min before emersion in fixative (1:3 acetic acid:methanol). A few follicles were selected per microscope slide, and slides were dried before staining with a 5% Giemsa staining solution for 15 min (5 mL Giemsa in 95 mL sodium-phosphate buffer, 6.8 pH). When dry, coverslips were mounted with DPX and slides were scanned systematically under a Leica DM E light microscope. Mitotic cells of suitable stage and clarity were selected for counting and analysis. Photographs were taken of metaphase or anaphase chromosome spreads at 1200× magnification using an Olympus BX51 Compound Microscope. CellSense Dimension 1.6 software (Olympus) was used to measure chromosomes. Each chromosome arm was measured separately and combined to give total chromosome length. For each mitotic spread measured, individual chromosomes were then ranked and numbered from longest to shortest. This information provided data for arm-length ratios for the nomenclature developed by Levan et al. (Citation1964). Because chromosome contraction is expected to result in variation among cells at different stages of cell division, the ratio of each chromosome to the total chromosome material per cell spread was calculated to allow comparison of relative chromosome sizes among cells and among individual wētā. Diploid species are expected to have pairs of homologous chromosomes that do not differ from one another significantly in length. A two-sample t-test implemented in Minitab 16 Statistical Software (Minitab Inc.) was used to compare the ranked individual chromosome lengths to identify likely chromosome pairings. This method allowed for inferences to be drawn about possible pairs by comparing the P-values of neighbouring chromosome pairs, although this method would only allow comparisons with a relatively small data set, as was used here. With larger samples, the P-values would decrease to zero and all chromosomes would be significantly differentiated because of being ranked by size before the t-tests were performed. As the pairs and sex chromosomes have only putatively been identified using visual methods in the past (e.g. Morgan-Richards Citation1997), we developed this approach to provide an unbiased method for identification of the unpaired sex chromosome.
Results
The karyotypes from the two individual wētā from a single population (H. thoracica from Hawkes Bay) were compared. Each chromosome in 10 mitotic cells was measured and variation between individuals was assessed visually using p-arm by q-arm scatter plots of all chromosomes (). Almost complete overlap was observed for the range of chromosome shape and relative size in the two wētā karyotypes. Only one pair of their smaller chromosomes can be visually identified on the graph (Chromosome 10 and 11, ). The autosomes can be divided into a cluster of larger metacentric chromosomes and a cluster of small pairs. Two of the smaller chromosomes (10th and 11th) have two arms but the smaller arm (p-arm) is only visible (and measurable) when mitosis has proceeded to only partial chromosome contraction. Full chromosome contraction results in mitotic cells where these chromosomes appear to have a single arm. Hence, for both individual wētā, two clusters of these chromosomes are resolved on the graph resulting from variation in mitotic stage and hence visibility or not of the p-arm. After confirming that individual cells from the same population have similar karyotypes, one mitotic spread from each specimen (n = 9, n = 10, n = 10, n = 13) from each of the four populations was measured and analysed. The wētā from the two H. thoracica populations have very similar karyotypes (–, ). Both have nine large metacentric chromosomes that vary in relative size by only a small amount. Two smaller submetacentric chromosomes (10 and 11) were identified in the Hawkes Bay group of H. thoracica, although as the centromeres were only visible in about half of the spreads measured, it is likely that centromeres were not visible in the Manawatu group because of greater chromosome constriction, as they have a similar total length and are probably homologous. The last six chromosomes in each of the two H. thoracica populations were very small, and no centromere could be identified. They were labelled telocentric, and the ratio was given based on an arbitrary value of 0.1% for the p-arm. Chromosome nine was significantly differentiated in the Hawkes Bay sample and in the first individual wētā studied. For the Manawatu H. thoracica and the second individual wētā, no chromosome stood out as being significantly differentiated from the rest, so chromosome nine was considered to be the most likely sex chromosome candidate for H. thoracica (; –).
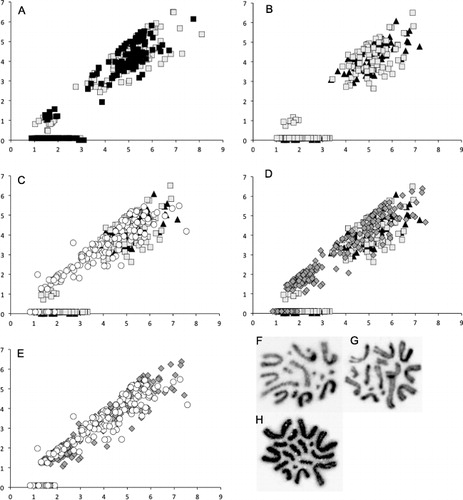
Table 1 Average relative length of each chromosome in the karyotype of Hawkes Bay male Hemideina thoracica (n = 10), and the average ratio for arm lengths (q/p) provides centromere position, and nomenclature based on Levan et al. (Citation1964). Metacentric (m 1.05–1.66); submetacentric (sm 1.67–2.99); subtelocentric (st 3.00–6.99); telocentric (t 7.00–39.00). The P-value used to assign chromosome pairs was taken from two-sample t-tests for differences between chromosomes.
Table 2 Average relative length of each chromosome in the karyotype of Manawatu male Hemideina thoracica (n = 10), and the average ratio for arm lengths (q/p) provides centromere position, and nomenclature based on Levan et al. (Citation1964). Metacentric (m 1.05–1.66); submetacentric (sm 1.67–2.99); subtelocentirc (st 3.00–6.99); telocentric (t 7.00–39.00). The P-value used to assign chromosome pairs was taken from two-sample t-tests for differences between chromosomes.
Hemideina crassidens has a distinct karyotype. This species differed in total number of chromosomes, having 15 chromosomes rather than 17 (diploid number in males). There are 11 relatively large metacentric chromosomes that showed a greater range of size than the large chromosomes in the H. thoracica karyotype (; ), although the largest chromosomes of H. crassidens are homologous in size (proportion of the genome) with the same centromeric position to those of H. thoracica. Hemideina crassidens has two intermediate-sized metacentric chromosomes that appeared to be a homologous pair. They are larger than the smaller well-differentiated pair belonging to H. thoracica, although the smallest pair of H. crassidens chromosomes appears to be very similar in size to the three smallest pairs of the H. thoracica set. When individual chromosomes were compared statistically using a two-sample t-test (), the fifth chromosome was shown to be most significantly differentiated from its two neighbouring chromosomes, and is therefore most likely to be the sex chromosome ().
Table 3 Average relative length of each chromosome in the karyotype of Manawatu male Hemideina crassidens (n = 10), and the average ratio for arm lengths (q/p) provides centromere position, and nomenclature based on Levan et al. (Citation1964). Metacentric (m 1.05–1.66); submetacentric (sm 1.67–2.99); subtelocentirc (st = 3.00–6.99); telocentric (t 7.00–39.00). The P-value used to assign chromosome pairs was taken from two-sample t-tests for differences between chromosomes.
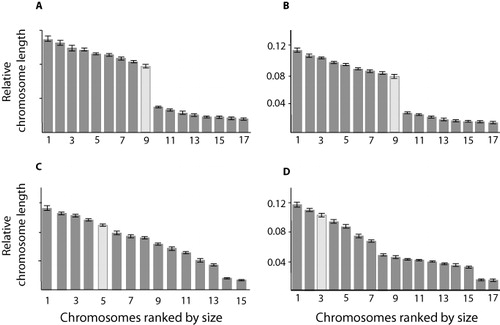
Hemideina trewicki also has a distinct karyotype, with only seven large metacentric chromosomes, that roughly matched the relative size of the largest chromosomes from H. thoracica and H. crassidens (; ). There appeared to be four pairs of chromosomes that all roughly correspond to the size and centromeric position of the intermediate-sized (second-smallest) chromosome pair of H. crassidens. The smallest pair matches the smallest chromosomes from both H. crassidens and H. thoracica. The sixth and seventh chromosomes appeared to make a pair when examined visually, and the two-sample t-test values support this inference; however, as these chromosomes overlap in size with the larger H. thoracica and H. crassidens chromosomes they are not distinguishable among species. Overall, despite differing numbers of chromosomes, H. crassidens and H. trewicki appear to have a more similar karyotype than either have when compared with H. thoracica. A two-sample t-test gave the most significantly differentiated size for the third chromosome (), so this is probably the sex chromosome for H. trewicki ().
Table 4 Average relative length of each chromosome in the karyotype of Hawkes Bay male Hemideina trewicki (n = 9), and the average ratio for arm lengths (q/p) provides centromere position, and nomenclature based on Levan et al. (Citation1964). Metacentric (m 1.05–1.66); submetacentric (sm 1.67–2.99); subtelocentirc (st 3.00–6.99); telocentric (t 7.00–39.00). The P-value used to assign chromosome pairs was taken from two-sample t-tests for differences between chromosomes.
Discussion
The two populations of H. thoracica from Hawkes Bay and Manawatu belong to the same chromosome race, and all three species examined here have distinct karyotypes. The H. thoracica karyotype has the only autosome pair that is well-differentiated from all other chromosomes in the complement (chromosome 10 and 11; pair 5). Hemideina crassidens and H. trewicki have chromosomes that are not shared with H. thoracica. Both H. thoracica and H. crassidens have unique chromosomes that can be used to distinguish them as a species/chromosome race. Hemideina thoracica and H. trewicki also have quantitative differences in the number of certain sized chromosomes (three very small pairs in H. thoracica as opposed to one in both other species, and four intermediate sized pairs between 3.11 and 4.74 units (see ratio column in and ) in H. trewicki as opposed to one in H. crassidens and none in H. thoracica). These differences may be important in distinguishing the ancestry of chromosomes in the karyotype of putative hybrids.
Previous cytogenetic studies that aimed to compare hybrid zones in Hemideina spp. have labelled the longest chromosome in the karyotype as the sex chromosome (e.g. Morgan-Richards Citation1997). As all chromosomes are paired in males except the sex chromosome, the odd number of large chromosomes in each karyotype implies that one is unpaired and therefore the sex chromosome. However, as all chromosomes vary only slightly in size it was impossible to accurately visually identify one solitary chromosome that did not match any other in most Hemideina karyotypes. It is possible that the sex chromosome is identical in size to one of the autosomal pairs, although the results of the detailed measurements and comparison of relative chromosome length after ranking revealed that for H. crassidens, the fifth chromosome is most probably the sex chromosome (, ), whereas in H. trewicki the most likely sex chromosome is the third-largest metacentric (, ). No single chromosome was unequivocally un-paired within the H. thoracica karyotype; however, the ninth chromosome seems the most likely candidate for the sex chromosome (, –). The centromere positions that were calculated (–) did not help differentiate the sex chromosome from the autosome pairs. The sex chromosomes in all three species are most likely homologous, but extensive rearrangements in autosomes will result in different relative lengths of the autosomes to the sex chromosome.
As previous studies (e.g. Trewick & Morgan-Richards Citation2005; Bulgarella et al. Citation2014) have shown that H. crassidens and H. trewicki are genetically more similar than either is to H. thoracica, the closer similarities of karyotypes is expected. They are closer in relative chromosome sizes than either are to H. thoracica, and it is also possible that one set of extra chromosomes in H. trewicki is due to the fission or fusion of the chromosome pair that belong exclusively to H. crassidens, which could explain its absence in the karyotypes of the other species. Due to the position of the sex chromosomes, it appears that in H. trewicki, one of the largest two autosomal pairs present in H. crassidens also probably went through the process of fission or fusion, to give two of the intermediate-sized pairs found in H. trewicki but not in H. crassidens. Due to the larger number of rearrangements between H. thoracica and the other two species, it is difficult to draw any inferences from the size of the sex chromosome. The differences between karyotypes also imply that hybrids between H. crassidens and H. trewicki have a higher chance of being fertile than hybrids between H. thoracica and the other two species, because of problems with chromosome alignments and recombination in F1 progeny, although this is yet to be determined.
The centromere position for all chromosomes (which were large enough for the centromere to be identified) was metacentric (). Even though this applied to all three species it would be wrong to infer some kind of karyotypic constraint in this lineage. There are many other chromosome races within Hemideina that do not conform to this pattern (e.g. the 23/24 and another 17/18 chromosome race of H. thoracica (Morgan-Richards & Wallis Citation2003), as well as the 19/20 chromosome race of H. crassidens (Morgan-Richards Citation2000)), so this is likely to be the result of chance. Tree and giant wētā (genera Hemideina and Deinacrida) do appear to have a high tolerance for chromosome rearrangements, as is evident from the large number of distinct karyotypes and hybrid zones within three species. The variation seen in Hemideina appears to be the result of Robertsonian translocations, rather than duplication or loss of genetic material, as Morgan-Richards (Citation2005) showed that genome size was similar among different chromosome races of H. thoracica.
Different evolutionary lineages differ greatly in the extent of chromosome rearrangements that have been maintained (e.g. Bourque et al. Citation2005 and references therein). Extensive chromosome rearrangements have been linked to speciation and adaptive radiations in some lineages, e.g. Onychophora: Peripatopsidae (Rowell et al. Citation1995, Citation2002) but many mammalian chromosome races show no phenotypic differentiation (Searle Citation1993). In conclusion, this study shows that the three Hemideina species in narrow regions of sympatry have distinct karyotypes, which can be used in future to identify putative hybrids. The sex chromosomes for each species were tentatively identified using the more rigorous measurements presented here, and comparisons between the three species’ karyotypes match previous inferences about relatedness based on DNA sequences.
Associate Editor: Dr Rob Cruickshank.
Acknowledgements
Paul Barrett helped with microscope and software. Mariana Bulgarella, Emily Koots, Rose Gregerson and Phoebe Donovan helped to collect wētā. Massey University provided funding (MURF: can wētā coexist?).
References
- Bourque G, Zdobnov EM, Bork P, Pevzner PA, Tesler G 2005. Comparative architectures of mammalian and chicken genomes reveal highly variable rates of genomic rearrangements across different lineages. Genome Research 15: 98–110.10.1101/gr.3002305
- Bulgarella M, Trewick S, Minards NA, Jacobson MJ, Morgan-Richards M 2014. Shifting ranges of two tree weta species (Hemideina spp.): competitive exclusion and changing climate. Journal of Biogeography. 41: 524–535. 10.1111/jbi.12224
- Griffin MJ, Trewick SA, Wehi PM, Morgan-Richards M 2011. Exploring the concept of niche convergence in a land without rodents: the case of weta as small mammals. New Zealand Journal of Ecology 35: 302–307.
- Kirkpatrick M 2010. How and why chromosome inversions evolve. PLoS Biology 8: e1000501. 10.1371/journal.pbio.1000501.g002
- Kirkpatrick M, Barton N 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434.10.1534/genetics.105.047985
- Levan A, Fredga K, Sandberg AA 1964. Nomenclature for centromeric position on chromosomes. Hereditas 52: 201–220.10.1111/j.1601-5223.1964.tb01953.x
- Morgan-Richards M 1997. Intraspecific karyotype variation is not concordant with allozyme variation in the Auckland tree weta of New Zealand, Hemideina thoracica (Orthoptera: Stenopelmatidae). Biological Journal of the Linnean Society 60: 423–442.
- Morgan-Richards M 2000. Robertsonian translocations and B chromosomes in the Wellington tree weta, Hemideina Crassidens (Orthoptera: Anostostomatidae). Hereditas 132: 49–54.10.1111/j.1601-5223.2000.00049.x
- Morgan-Richards M 2005. Chromosome rearrangements are not accompanied by expected genome size change in the tree weta Hemideina thoracica (Orthoptera, Anostostomatidae). Journal of Orthoptera Research 14: 143–148.10.1665/1082-6467(2005)14[143:CRANAB]2.0.CO;2
- Morgan-Richards M, Daugherty CH, Gibbs GW 1995. Taxonomic status of tree weta from Stephens Island, Mt Holdsworth and Mt Arthur, based on allozyme variation. Journal of the Royal Society of New Zealand 25: 301–312.10.1080/03014223.1995.9517491
- Morgan-Richards M, Gibbs GW 2001. A phylogenetic analysis of New Zealand giant and tree weta (Orthoptera: Anostostomatidae: Deinacrida and Hemideina) using morphological and genetic characters. Invertebrate Systematics 15: 1–12.10.1071/IT99022
- Morgan-Richards M, King T, Trewick SA 2001a. The evolutionary history of tree weta: a genetical approach. In: Field L ed. Biology of weta, king crickets and their Allies. Oxford, CABI Publishing. Pp. 111–124.
- Morgan-Richards M, Smissen RD, Shepherd LD, Wallis GP, Hayward JJ, Chan C-H et al. 2009. A review of genetic analyses of hybridisation in New Zealand. Journal of the Royal Society of New Zealand 39(1): 15–34.10.1080/03014220909510561
- Morgan-Richards M, Townsend JA 1995. Hybridisation of tree weta on Banks Peninsula, New Zealand, and colour polymorphism within Hemideina ricta (Orthoptera: Stenopelmatidae). New Zealand Journal of Zoology 22: 393–399.10.1080/03014223.1995.9518058
- Morgan-Richards M, Trewick SA, Wallis GP 2000. Characterization of a hybrid zone between two chromosomal races of the weta Hemideina thoracica following a geologically recent volcanic eruption. Heredity 85: 586–592.10.1046/j.1365-2540.2000.00796.x
- Morgan-Richards M, Trewick SA, Wallis GP 2001b. Chromosome races with Pliocene origins: evidence from mtDNA. Heredity 86: 303–312.10.1046/j.1365-2540.2001.00828.x
- Morgan-Richards M, Wallis GP 2003. A comparison of five hybrid zones of the weta Hemideina thoracica (Orthoptera: Anostostomatidae): degree of cytogenetic variation fails to predict zone width. Evolution 57: 849–861.10.1111/j.0014-3820.2003.tb00296.x
- Ramsay GW, Bigelow RS 1978. New Zealand wetas of the genus Hemideina. The Weta 1: 32–34.
- Rieseberg LH 2001. Chromosomal rearrangements and speciation. Trends in Ecology & Evolution 16: 351–358.10.1016/S0169-5347(01)02187-5
- Rowell DM, Higgins AV, Briscoe DA, Tait NN 1995. The use of chromosomal data in the systematics of viviparous onychophorans from Australia (Onychophora: Peripatopsidae). Zoological Journal of the Linnean Society 114: 139–153.10.1111/j.1096-3642.1995.tb00117.x
- Rowell DM, Rockman MV, Tait NN 2002. Extensive Robertsonian rearrangement: implications for the radiation and biogeography of Planipapillus Reid (Onychophora: Peripatopsidae). Journal of Zoology 257: 171–179.10.1017/S0952836902000778
- Searle JB 1993. Chromosome hybrid zones in eutherian mammals. In: Harrison RG ed. Hybrid zones and the evolutionary process. New York, NY, Oxford University Press. Pp. 309–353.
- Sessions SK 1996. Chromosomes: molecular systematics. In: Hillis DM, Moritz C eds. Molecular systematics. 2nd edition. Sunderland, MA, Sinauer Associates. Pp. 121–168.
- Sites JW, Moritz C 1987. Chromosomal evolution and speciation revisited. Systematic Biology 36: 153–174.
- Trewick SA, Morgan-Richards M 2005. After the deluge: mitochondrial DNA indicates Miocene radiation and Pliocene adaptation of tree and giant weta (Orthoptera: Anostostomatidae) Molecular phylogenetics of weta. Journal of Biogeography 32: 295–309.10.1111/j.1365-2699.2004.01179.x
