ABSTRACT
Dietary analysis of nocturnal aerial insectivores such as bats is not possible by direct visual observation of predation, therefore diet is traditionally determined by analysing dietary remains in scats. Analysing DNA in scats through metabarcoding can reveal dietary items not preserved in faeces but also resolve dietary preferences to species level. DNA metabarcoding analysis of scats collected from six individual long-tailed bats (Chalinolobus tuberculatus) from Whirinaki Forest Park revealed a predominance of aquatic insects (Ephemeroptera and Trichoptera) and small-bodied moths (Lepidoptera). This contrasts with earlier studies of faecal remains in this species which revealed a strong preference for Dipterans and Coleopterans. The use of DNA metabarcoding may provide a better understanding of diets, as soft-bodied insects can be detected even when not apparent in physical examinations of scat, and thus will allow for more robust comparison of differences in diets between populations and seasons. The application of DNA metabarcoding is a simple but high-resolution tool to allow further study on dietary preference in this endangered species.
Introduction
Molecular scatology, or the analysis of faecal DNA, has become an increasingly common technique in animal ecology. Faecal DNA can be used for species confirmation and phylogenetic analyses (Guan et al. Citation2020) to identify individuals in studies of population size and home range (Treloar et al. Citation2023), and increasingly in studies of diet (Thuo et al. Citation2019). The technique is particularly useful when direct observation of feeding is not possible, especially for species that are small, nocturnal and volant, and where prey remains are not identifiable in faeces. The use of faecal DNA metabarcoding to study animal diet has increased significantly in recent years (Ando et al. Citation2020) and has been applied to study the diets of insectivorous bats (Bohmann et al. Citation2011; Clare et al. Citation2011; Burgar et al. Citation2014; Alberdi et al. Citation2020; Ingala et al. Citation2021).
The New Zealand long-tailed bat (LTB; Chalinolobus tuberculatus) is a typical microbat of the family Vespertilionidae, being a small-bodied (∼10 g body weight), fast flying (∼60 km/h), aerial insectivore and exclusively nocturnal (O'Donnell Citation2001). This species is classified as nationally critical by the New Zealand Department of Conservation (O’Donnell et al. Citation2017). Microbat diet is clearly amenable to study by faecal metabarcoding because direct observations of feeding are impossible and small insect prey may be too thoroughly digested to provide identifiable faecal remains. Previous studies of LTB diet have been undertaken by visually identifying prey remains in scats collected from captured bats or from below roosts. Gillingham (Citation1996) found that the diet was dominated by Diptera, followed by Coleoptera and Lepidoptera. Aquatic taxa such as Trichoptera and Ephemeroptera were very minor components of the diet (Gillingham Citation1996). Similar results were obtained by Gurau (Citation2014) who found the diet to be dominated by Diptera, followed by Lepidoptera and Coleoptera, with Trichoptera, Hymenoptera and mites comprising extremely small proportions of the diet. Long-tailed bats often feed over or near water (Griffiths Citation2007) so it is unsurprising that aquatic dipterans, such as mosquitoes and chironomid midges, are common prey, but strange that other aquatic insects, such as Ephemeroptera, Plecoptera and Trichoptera do not feature more prominently in their diet (Gurau Citation2014). Both Gillingham (Citation1996) and Gurau (Citation2014) noted that much of the prey remains were unidentifiable and it is entirely possible that softer bodied insects are either too thoroughly masticated or digested to provide identifiable faecal remains. Metabarcoding DNA analysis of scats not only allows for identification of prey that may leave no visible trace in faeces but can also provide identification of prey to species level so long as sequence data are available for all likely prey taxa. Metabarcoding is increasingly used to analyse eDNA in New Zealand freshwaters and a large number of terrestrial and freshwater invertebrate taxa now have metabarcoding sequence data available. This study represents a preliminary analysis of faecal DNA from LTB scat samples to investigate dietary preference in this endangered species.
Methods
Scat samples were collected from long-tailed bats captured in Whirinaki Forest Park (38.6448° S, 176.7073° E) in November 2022 as part of a tagging programme organised by the New Zealand Department of Conservation. All bat samples analysed here were recovered from bats caught in a single night to avoid the likelihood of recapturing bats, and assumptions on diet being biased by any one individual. Scat samples were opportunistically recovered from cloth bags used to hold individual bats awaiting tagging or release. One scat sample from each of six individual bats was preserved in 95% ethanol and later transferred to DNA/RNA shield medium (Zymo Research, USA) prior to analysis by Wilderlab NZ Ltd (Miramar, Wellington).
DNA quality analysis and NGS library preparation
DNA quality/quantity analysis, adapter-fusion, indexing and amplification were carried out in single-step quantitative PCR reactions on an Applied Biosystems QuantStudio 1 qPCR instrument. DNA extracts were PCR-amplified using the Wilderlab comprehensive multi-species analysis package (). Fusion-tagged forward primers were designed as follows: P5 Illumina adapter sequence + Illumina TruSeq sequencing primer + 8-bp unique index tag + locus specific forward primer and fusion-tagged reverse primers as: P7 Illumina adapter sequence + 8-bp unique index tag + locus specific reverse primer. All 8-bp index tags differed from each other by at least 3 bp. Each reaction contained 5 µl SensiFAST 2x LoRox SYBR Mix (Bioline), 0.25 µl forward primer (10 µM), 0.25 µl reverse primer (10 µM), 0.5 µl BSA (10 mg ml-1, Sigma Aldrich), 2 µl de-ionised water and 2 µl template DNA. qPCR cycling conditions included an initial denaturation of 3 min at 95°C; followed by 40 cycles of 5 s at 95°C, 10 s at 52°C, 15 s at 72°C. DNA quality and quantity were assessed by checking that a sigmoidal log-amplification curve was visible with a CT value < 35. A negative control reaction containing 2 µl de-ionised water in place of the template DNA was included with each run. NGS libraries were pooled at roughly equimolar concentration using the final normalised ΔRn fluorescence values as a guide and cleaned and double-end size selected using AMPure XP magnetic beads (0.9× and 1.2× for lower and upper size bounds, respectively). The final pooled library concentration was determined using a Qubit 4 fluorometer (ThermoFisher Scientific) and the concentration was adjusted to 50 pM in sterile DNAse/RNAse free water. The library was then loaded onto an iSeq i1 V2 reagent cartridge with 300-cycle flow cell (Illumina) with 5% Phi X and run for 290 cycles in a single direction on an Illumina iSeq 100 instrument.
Table 1. Locus-specific sections of the fusion-tag metabarcoding primers used in the Wilderlab basic freshwater panel, Jan 2023, with references where applicable.
ASV generation and taxonomic assignment
The iSeq 100 output sequence fastq file was de-multiplexed in R (R Core Team, Citation2021) using the insect package (v 1.4.0; (Wilkinson et al. Citation2018)) and trimmed sequences were filtered to produce a table of exact amplicon sequence variants (ASVs) using the DADA2 R package (Callahan et al. Citation2016). ASVs were identified to the lowest possible taxonomic rank using Wilderlab’s four-step classification process. This involves: (1) exact matching against an intensively-curated database of previously detected eDNA sequences; (2) remaining (i.e. non-matched) sequences are exact-matched against a global reference sequence database primarily compiled of trimmed reference sequences downloaded from GenBank and the Barcode of Life data System (BOLD) then matching sequences assigned at the lowest common ancestor level (LCA; i.e. assigned to genus level if matched with 100% identity to more than one species, or to family level if matched to more than one genus); (3) remaining sequences that are >50 bp in length are matched with single indel/substitution tolerance against the same GenBank/BOLD reference database and matching sequences assigned at LCA level; and (4) remaining sequences are queried against the local GenBank/BOLD reference database using the SINTAX classification algorithm (Edgar Citation2016) with a conservative assignment threshold of >0.99 and maximum genus level assignment threshold.
Results
In all samples, sequence read abundance was greatest for long-tailed bat DNA which varied from 1866 to 37,369 reads. Sequence reads for prey taxa were therefore normalised by dividing by the read abundance for LTB as one way to account for total DNA abundance. Of the scat samples collected from six individual bats, one sample did not provide any useful DNA metabarcoding sequences that could be identified below the highest taxonomic resolution of insects and spiders. However, the other five samples provided recoverable DNA identifiable to at least family and, in many cases, species level (). The sixth individual provided DNA only identifiable to the higher-level taxa of insects and spiders. The total number of dietary taxa identified to family level or below was 24 and the number of taxa varied from 3 to 12 in each individual scat sample. Aquatic insects dominated the diet in these bats, especially species of Ephemeroptera and Trichoptera, with all five bats that provided recoverable DNA having consumed these taxa. Total DNA reads for these groups comprising 62% of all dietary DNA. Small-bodied lepidopterans also featured prominently: brown-headed leafroller moth (Ctenopseustis fraterna), green-headed leafroller moth (Planotortrix excessana), grass moth (Culladia cuneiferellus), filata moth (Chloroclystis filata). Lacewings (Micromus tasmaniae) were consumed by three individuals. One bat had consumed a species of longhorn beetle (Arhopalus sp.), while another contained DNA from a spider (Clubiona consensa) and a weevil (Curculionidae) indicating feeding on non-volant prey, although the latter may have been captured in flight given that most weevil species can fly. Spider DNA was also identified in one other individual.
Figure 1. Normalised metabarcoding sequence reads for all dietary taxa identified to family level or below. Symbols represent aquatic volant taxa (asterisks), terrestrial volant taxa (open circles) and non-volant terrestrial taxa (open diamond). Horizontal bars are the geometric mean of data.
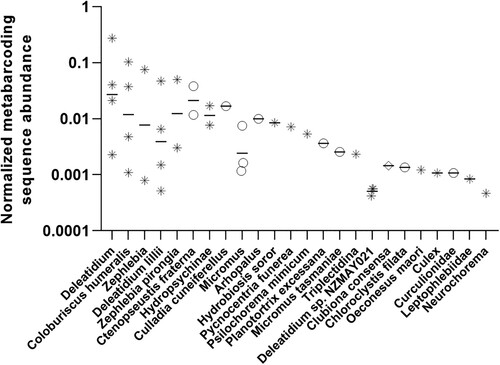
Inferring dietary dominance from metabarcoding read sequence abundance is notoriously problematic and several approaches to analysing such data have been proposed (Deagle et al. Citation2019). Using the recommended relative read abundance (RRA) approach of Deagle et al. provides a slightly different interpretation of the relative importance of some of the dietary groups identified when higher-level taxa are included; for instance, a relatively large number of lepidopteran reads were not identified at lower taxonomic levels. However, it must be acknowledged that the same dietary item may account for reads at both the species and higher taxonomic levels. Data for individuals using the RRA method are given in . Overall, 36.4% of DNA reads were unable to be assigned to any taxon lower than the level of insects and could represent species from any of these insect orders.
Table 2. Dietary taxa in scat samples from six long-tailed bats quantified by relative read abundance (%).
Discussion
The most striking contrast between these molecular data and previous studies of long-tailed bat diet undertaken by identification of physical remains in scats is the dominance of relatively large-bodied aquatic taxa of mayflies (Ephemeroptera) and caddis flies (Trichoptera). This is not unexpected given the common observation that LTBs often forage over or near water (Griffiths Citation2007; Dekrout et al. Citation2014). It is possible that these soft-bodied insects are too thoroughly masticated or digested to be identifiable in scats and both the studies of Gillingham (Citation1996) and Gurau (Citation2014) found that a large number of insect remains were unidentifiable. It is also possible that these taxa did not feature in the previous studies because those samples were obtained from bats feeding away from water or due to seasonal variation in the abundance of aerial insect prey. Terrestrial insect taxa also featured prominently in the molecular data although the strong preference for coleopterans identified in previous studies was not evident in our samples. Hard-bodied coleopterans may more readily survive digestion and therefore feature more prominently than is actually the case. Given the cost in time to analyse scats by physical observation, molecular analysis is a powerful tool to investigate seasonal and geographic variability in LTB diet, and can identify multiple prey items in a single scat and often to species level if the relevant sequence data are available. The number of species for which such data are known is rapidly increasing. However, a significant proportion of DNA reads were assigned to the higher taxa of spiders and insects, most notably in LTB6 where no lower-level taxa were identified. The most likely reason for this is the quality of DNA which varies considerably due to factors such as sample preservation (Panasci et al. Citation2011), time since consumption (Thuo et al. Citation2019) and digestive efficiency which can vary considerably between different bat prey (Barclay et al. Citation1991).
Mayflies and caddis flies are key components of the three major groups of pollution sensitive taxa, the EPT taxa (Ephemeroptera, Plecoptera, Trichoptera) distinctive of good water quality and which dominate the macroinvertebrate community index assessment for water quality in New Zealand (Stark and Maxted Citation2007). If these pollution sensitive taxa are crucially important prey for long-tailed bats, then degrading water quality in lowland and urban streams potentially reduces preferred prey availability. However, the ability of LTBs to forage opportunistically on whatever aerial insect prey are available may offset such losses, although terrestrial insect abundance is generally in decline in many parts of the world due to factors including land use changes, pesticide use and climate change (Møller Citation2020). There is currently little understanding of the status and trend of terrestrial insects in New Zealand and this has been identified as a major knowledge gap (Barnsley Citation2021). Our molecular data did support the previous observation of Gurau (Citation2014), who identified remains of lepidopteran larvae in scats, that LTBs may opportunistically prey on non-volant prey, although eDNA is unable to distinguish between volant adults and non-volant larvae.
Acknowledgements
We wish to thank the Department of Conservation for supporting this research through the opportunity for TS to attend the Whirinaki bat tagging programme and obtain scat samples for analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Alberdi A, Razgouor O, Aizpurua O, Novella-Fernandez R, Aihartza J, Budinski I, Garin I, Ibanez C, Izagirre E, Rebelo H, et al. 2020. DNA metabarcoding and spatial modelling link diet diversification with distribution homogeneity in European bats. Nature Communications. 11:1154. doi:10.1038/s41467-020-14961-2.
- Ando H, Mukai H, Komura T, Dewi T, Ando M, Isagi Y. 2020. Methodological trends and perspectives of animal dietary studies by non-invasive fecal DNA metabarcoding. Environmental DNA. 2:391–406. doi:10.1002/edn3.117.
- Barclay RMR, Dolan MA, Dyck A. 1991. The digestive efficiency of insectivorous bats. Canadian Journal of Zoology. 69:1853–1856. doi:10.1139/z91-256.
- Barnsley J. 2021. What is known about terrestrial insect population trends in Aotearoa New Zealand? Intern Project – Peer-reviewed report: Office of the Prime Minister’s Chief Science Advisor, Wellington. 31 p.
- Bohmann K, Monadjem A, Lehmkuhl Noer C, Rasmussen M, Zeale MRK, Clare E, Jones G, Willerslev E, Gilbert MTP. 2011. Molecular diet analysis of two African free-tailed bats (Molossidae) using high throughput sequencing. PLoS One. 6:e21441. doi:10.1371/journal.pone.0021441.
- Burgar JM, Murray DC, Craig MD, Haile J, Houston J, Stokes V, Bunce M. 2014. Who’s for dinner? High-throughput sequencing reveals bat dietary differentiation in a biodiversity hotspot where prey taxonomy is largely undescribed. Molecular Ecology. 23:3605–3617. doi:10.1111/mec.12531.
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods. 13:581–583. doi:10.1038/nmeth.3869.
- Clare EL, Barber BR, Sweeney BW, Hebert PDN, Fenton MB. 2011. Eating local: influences of habitat on the diet of little brown bats (Myotis lucifugus). Molecular Ecology. 20:1772–1780. doi:10.1111/j.1365-294X.2011.05040.x.
- Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJ, Clare EL, Kartzinel TR, Eveson JP. 2019. Counting with DNA in metabarcoding studies: how should we convert sequence reads to dietary data? Molecular Ecology. 28:391–406. doi:10.1111/mec.14734.
- Dekrout A, Clarkson BD, Parsons S. 2014. Temporal and spatial distribution and habitat associations of an urban population of New Zealand long-tailed bats (Chalinolobus tuberculatus). New Zealand Journal of Zoology. 41:285–295. doi:10.1080/03014223.2014.953551.
- Edgar RC. 2016. SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv preprint: doi:10.1101/074161.
- Gillingham NJ. 1996. The behaviour and ecology of long-tailed bats (Chalinolobus tuberculatus Gray) in the central North Island [Master of Science thesis]. Palmerston North: Massey University.
- Griffiths RW. 2007. Activity patterns of long-tailed bats (Chalinolobus tuberculatus) in a rural landscape, South Canterbury, New Zealand. New Zealand Journal of Zoology. 34:247–258. doi:10.1080/03014220709510083.
- Guan X, Britzke ER, Piaggio AJ, Bergman DL, Van Pelt L, Lance RF. 2020. Genetic assays for guano identification of species and sex in bats of the United States and Canada. Journal of Mammalogy. 101:970–978. doi:10.1093/jmammal/gyaa059.
- Gurau AL. 2014. The diet of the New Zealand long-tailed bat, Chalinolobus tuberculatus [Master of Science thesis]. Massey University.
- Ingala MR, Simmons NB, Wultsch C, Krampis K, Provost KL, Perkin SL. 2021. Molecular diet analysis of neotropical bats based on fecal DNA metabarcoding. Ecology and Evolution. 11:7474–7491. doi:10.1002/ece3.7579.
- Miya M, Sato Y, Fukunaga T, Sado T, Poulsen JY, Sato K, Minamoto T, Yamamoto S, Yamanaka H, Araki H, et al. 2015. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. Royal Society Open Science. 2:150088. doi:10.1098/rsos.150088.
- Møller AP. 2020. Quantifying rapidly declining abundance of insects in Europe using a paired experimental design. Ecology and Evolution. 10:2446–2451. doi:10.1002/ece3.6070.
- Nester GM, De Brauwer M, Koziol A, West KM, DiBattista JD, White NE, Power M, Heydenrych MJ, Harvey E, Bunce M. 2020. Development and evaluation of fish eDNA metabarcoding assays facilitate the detection of cryptic seahorse taxa (family: Syngnathidae). Environmental DNA. 2:614–626. doi:10.1002/edn3.93.
- O'Donnell CFJ. 2001. Advances in New Zealand mammalogy 1990–2000: long-tailed bat. Journal of the Royal Society of New Zealand. 31:43–57. doi:10.1080/03014223.2001.9517638.
- O’Donnell CFJ, Borkin KM, Lloyd CB, Parsons S, Hitchmough RA. 2017. Conservation status of New Zealand bats, 2017. Wellington: Department of Conservation. 4 p.
- Panasci M, Ballard WB, Breck S, Rodriguez D, Densmore LD, Webster DB, Baker RJ. 2011. Evaluation of fecal DNA preservation techniques and effects of sample age and diet on genotyping success. The Journal of Wildlife Management. 75:1616–1624. doi:10.1002/jwmg.221.
- R Core Team. 2021. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/.
- Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E. 2011. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Research. 39:e145. doi:10.1093/nar/gkr732.
- Stark JD, Maxted JR. 2007. A user guide for the Macroinvertebrate Community Index. Prepared for the Ministry for the Environment. Cawthron Report No.1166. 58 p.
- Thuo D, Furlan E, Broekhuis F, Kamau J, MacDonald K, Gleeson D. 2019. Food from faeces: evaluating the efficacy of scat DNA metabarcoding in dietary analyses. PLos One. 14:e0225805. doi:10.1371/journal.pone.0225805.
- Treloar S, Lohr C, Hopkins AJM, Ottewell K, McArthur S, Davis RA. 2023. Scat DNA as a non-invasive method for estimating the abundance of the vulnerable mala (Lagorchetes hirsutus). Wildlife Research. doi:10.1071/WR22122.
- Vamos EE, Elbrecht V, Leese F. 2017. Short COI markers for freshwater macroinvertebrate metabarcoding. Metabarcoding and Metagenomics. 1:e14625. doi:10.3897/mbmg.1.14625.
- Wilkinson SP, Davy SK, Bunce M, Stat M. 2018. Taxonomic identification of environmental DNA with informatic sequence classification trees. PeerJ preprint: doi:10.7287/peerj.preprints.26812v1.
