Abstract
Background
The Lisu group is a unique minority in Yunnan province. However, there is a lack of Y-STR population data for Chinese Lisu and the genetic structure of the Lisu group and other populations is unclear.
Aim
To provide genetic data for 23 Y-STRs in the Chinese Lisu population from Chuxiong Yi Autonomous Prefecture, as well as to analyse population genetic relationships between Chinese Lisu ethnic minority and other reference groups.
Subjects and methods
423 unrelated healthy Lisu males were genotyped using the PowerPlex® Y23 system. Forensic parameters were calculated according to the previously published studies. Genetic structure analysis among Chinese Lisu and other populations was conducted using the YHRD’s AMOVA tools.
Results
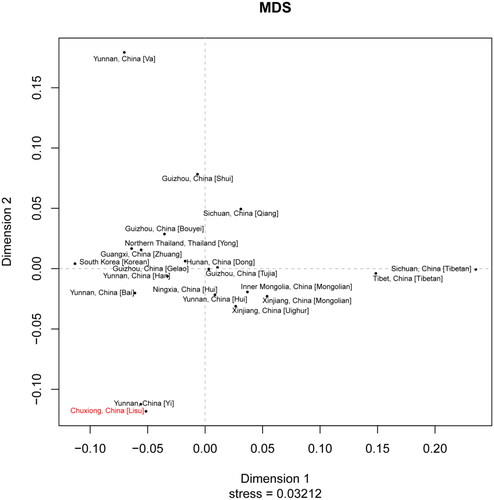
Gene diversity (GD) ranged from 0.2,466 (DYS438) to 0.8,945 (DYS385a/b) among the 23 Y-STR loci. According to haplotype analysis, 323 different haplotypes were obtained, out of which 271 were unique. The haplotype diversity (HD) and discrimination capacity (DC) were 0.9,977 and 0.7,636, respectively. MDS plot indicated that the Chuxiong Lisu group is genetically related to the Yunnan Yi group.
Conclusions
This is the first report on Y-STR population data for the Chinese Lisu population. These data would be valuable for forensic applications.
Introduction
The Y chromosome is male-specific, paternally inherited, and haploid for the majority of its length because it is immune to meiotic recombination. This region is passed down from father to son and remains unchanged unless a mutation event occurs. As a result, the Y chromosome contains a record of all the mutational events that occurred among his ancestors, reflecting the history of his paternal lineage. It is possible to reconstruct the histories of paternal lineages by comparing Y-STRs DNA polymorphism (Jobling and Tyler-Smith Citation1995). The interest in Y chromosome polymorphisms has steadily increased over the last few decades, not only because of its application in human and evolutionary genetics, but also because of its interest in forensics, particularly in cases where standard autosomal DNA profiling is not informative. Population data on Y-STRs is essential for crime scene investigations. Haplotypes composed of Y-STRs are used to characterise the paternal lineages of unknown male trace donors, particularly in sexual assault cases involving mixed stains, when both males and females contributed to the same trace (Dekairelle and Hoste Citation2001; Purps et al. Citation2015; Yin et al. Citation2022). Moreover, Y-STR haplotype analysis is increasingly employed in male relative identification (Ballantyne et al. Citation2010; Adnan et al. Citation2016), kinship analysis, familial searching (Foster et al. Citation1998), inferring paternal bio-geographical ancestry (Phillips Citation2015), and also for the analysis of migration (Underhill and Kivisild Citation2007), settlement, or mating structure of human populations.
Recently, some Y-STR studies on various ethnic minorities have been reported. However, population genetic data on Y-STR loci of the Chuxiong Lisu population are lacking, and the genetic relationship between Lisu and other ethnic groups is unclear. In order to establish a database of the Chinese Lisu population living in Chuxiong Yi Autonomous Prefecture, Yunnan province, Southwest China, we used a PowerPlex® Y23 (Promega Corporation) kit, which includes 23 Y-chromosomal short tandem repeat (Y-STR) loci, namely, DYS576, DYS389I, DYS448, DYS389II, DYS19, DYS391, DYS481, DYS549, DYS533, DYS438, DYS437, DYS570, DYS635, DYS390, DYS439, DYS392, DYS643, DYS393, DYS458, DYS385a/b, DYS456, and Y-GATA-H4. In this study, we presented allele frequencies and haplotype distributions of 23 Y-STR loci and compared pairwise genetic distances with other populations.
The Lisu people are one of China’s oldest ethnic groups, mainly living in the northwest of Yunnan province at an average altitude of 2,000 to 3,000 metres, and most of them live in concentrated communities in Weixi and Nujiang Lisu Autonomous Prefecture. The rest are distributed in Lijiang, Baoshan, Diqing, Dehong, Dali, Chuxiong prefectures or counties in Yunnan, as well as Xichang, Muli, and Dechang counties in Sichuan. Lisu are a cross-border ethnic group found in Myanmar, Thailand, and India. It is the 22nd largest ethnic minority with a population of 762,996 (0.0,527%) according to the Seventh National Population Census in 2020. On the basis of historical records, the Lisu ethnic group migrated frequently from the southeast Tibet Plateau to Japan and other Southeast Asian countries via Sichuan and Yunnan (Cang Citation1997). The Lisu are descendants of the ancient Di-Qiang, the most powerful nomadic tribe in northwest China between 1,700 and 300 BC, and were later divided into several ethnic groups (Liu Citation2007; Wang Wg Citation2011). Lisu people have their own language (Lisu), which belongs to the Chinese-Tibetan language family (Yi branch of the Tibetan-Myanmese Language Group) (Lewis Citation2009). The geographical locations of Lisu people in this paper is shown in Figure S1.
Subjects & methods
Sample preparation
Blood samples from 423 unrelated healthy Lisu individuals living in the Chuxiong Yi Autonomous Prefecture of Yunnan province were collected after signing informed consent prior to participating in this study. This study was approved by the Medical Ethics Committee of Kunming Medical University (permit number: KMMU2020MEC013). DNA was extracted from FTA cards with the Chelex-100 method (Walsh et al. Citation1991).
PCR amplification and capillary electrophoresis
PCR for 23 Y-STR loci was carried out on a GeneAmp PCR system 9,700 (Applied Biosystems, USA) using the PowerPlex® Y23 (Promega Corporation) PCR amplification kit. Electrophoresis of fragments was performed on an ABI 3130XL Genetic Analyser (Applied Biosystems, USA) and subsequently analysed by GeneMapper ID-X v.1.5 software (Thermo Fisher Scientific, Waltham, MA, USA). The DNA typing and assignment of nomenclature were based on the ISFG recommendations (Bar et al. Citation1997; Lincoln Citation1997). During the entire amplification process, 2,800 M Control DNA and Nuclease-Free Water were used as the positive and negative controls, respectively. Our lab has also been accredited by the China National Accreditation Service for Conformity Assessment (CNAS).
Analysis of data
Allelic frequencies and haplotype frequencies were estimated by direct gene counting. Single-marker GD was calculated according to Nei with the formula GD = n(1-Σpi2)/(n-1), where n is the total number of samples, and pi is the relative frequency of the i-th allele (Clegg Citation1987). HD was calculated similarly to GD. The DC was defined as the ratio between the number of different haplotypes and the total number of haplotypes. Pairwise genetic distances (Rst) and associated P values among 21 populations were conducted by analysis of molecular variance (AMOVA) and visualised using two multidimensional scaling (MDS) plot via the YHRD website (http://www.yhrd.org/Analyse/AMOVA). For the purposes of Rst calculation and MDS analysis, multi-locus markers were disregarded except DYS385 and DYF387S1, and all haplotypes with duplications, micro variants, and null-alleles were removed from this analysis according to the algorithm of YRHD’s AMOVA tools. Our lab participated and passed the YHRD quality control. The haplotypes obtained in this study have been submitted to the YHRD, and the population accession number is YA005719.
Results and discussion
Forensic parameters of the 23 Y-STR loci system when applied to the Chuxiong Lisu population
We provide forensic population data for the Lisu minority from Chuxiong Yi Autonomous Prefecture, Yunnan province, Southwest China, for the first time using the PowerPlex® Y23 kit. The genotype data for 23 Y-STR loci are presented in Table S1. One intermediate allele, 12.2, was observed and highlighted in one individual at DYS392 (see Table S1). The allele frequencies and locus diversities of 23 Y-STR are shown in Table S2. In the study, 189 alleles were observed at 23 Y-STR loci, with the number of alleles at each locus ranging from 4 at (DYS389I, DYS549, DYS393 and Y-GATA-H4) to 54 at DYS385a/b, and corresponding allelic frequencies ranged from 0.0,024 (for alleles occurring only once) to 0.8,629 (allele10 at DYS438). The lowest gene diversity was observed at locus DYS438 with a value of 0.2,466 and the highest at the locus DYS385a/b with a value of 0.8,945. 18 of the 23 Y-STR loci were at high levels of genetic polymorphism with GD values greater than 0.5 (see Table S2). The haplotype distributions are listed in Table S3. A total of 323 different haplotypes were found in 423 individuals, out of which 271 (83.90%) were observed only once (unique haplotypes), 32 different haplotypes occurred twice, 10 different haplotypes appeared thrice, and 8 distinct haplotypes were observed more than three times in the 58 remaining samples. The overall HD was calculated as 0.9,977 with a DC of 0.7,636. Our results indicated that these 23 Y-STRs were highly genetically polymorphic in the Chuxiong Lisu group.
Genetic relationship between the Chuxiong Lisu population and reference populations in China or adjacent countries
The genetic distances (Rst) and associated P values for the pairwise relationships of 9,280 haplotypes between Chuxiong Lisu and 20 other reference populations from China or adjacent nations are shown in Table S4. The detailed information for each population is shown in Table S5. With the comparison analysis, Chuxiong Lisu was found to be significantly different from all other groups except Yunnan Yi (P value = 0.0,028). Based on Rst values, the shortest genetic distance observed from Chuxiong Lisu was Yunnan Yi (Rst = 0.0,075), while the greatest was found with Sichuan Tibetan (Rst = 0.3,626), followed by Yunnan Va (Rst = 0.3,030). Consistent results were also obtained in the MDS plot (). The MDS plot based on Rst values clearly indicated that Chuxiong Lisu and Yunnan Yi clustered together and located in the lower left of the third quadrant, suggesting their close genetic relationship, which was consistent with historical records claiming that the Lisu and Yi are descendants of the ancient Di-Qiang people (Liu Citation2007; Sun et al. Citation2013). Both the Lisu and Yi language belong to the Yi branch of the Tibetan-Myanmese Language Group of the Chinese-Tibetan language family, so these two populations have a higher genetic affinity than other populations. Moreover, based on the study of physical anthropology, Lisu people have similarities in the facial features with those of Yi people who reside in plateau areas (Liu and Yu Citation1990). Our previous report on genetic diversities of autosomal STR loci discovered that the Chuxiong Lisu population had a close genetic relationship with the Yunnan Yi population (Zhang et al. Citation2019), the male subjects in these two articles are identical. Another study of 15 autosomal STR loci also found Chuxiong Lisu and Chuxiong Yi to be related (from Yunnan province) (Huang et al. Citation2020). The Yunnan Va population positioned in the upper left of the second quadrant. Tibetans from Sichuan and Tibet gathered together and situated on the right side of the horizontal axis. The Korean or Thailand people from Southeast Asian countries were located on the left side of the horizontal axis and shared a distant genetic relationship with Yunnan Lisu and Yunnan Yi people. The findings revealed that the Lisu people of Chuxiong migrated from other Southeast Asian countries rather than Korea and Thailand. According to Huang’s analysis of autosomal STR loci, Lisu people from five different settlements in Yunnan province (Baoshan, Dehong, Shangri-La, Lijiang, and Nujiang) are clustered with Lisu immigrants from Burma, who are from Southeast Asian countries (Huang et al. Citation2020). The Lisu immigrants in Chuxiong may be different from those in other Lisu settlements in Yunnan province. Although the exact origins of the Lisu are unknown, some scholars believe that they are descended from the ancient Di-Qiang people, a plateau nomadic tribe that once inhabited the Qinghai-Tibetan Plateau and gave rise to numerous ethnic groups in southern and southwestern China. More population data on different genetic markers and Lisu settlements must be studied to determine the genetic relationship of Lisu people. Our findings showed that ethnically close populations had higher genetic affinity than other populations.
Conclusions
We firstly genotyped 23 Y-STR loci from 423 Chuxiong Lisu males and provided the relative forensic parameters. In addition, we evaluated the genetic relationships between the Chuxiong Lisu population and 20 other reference populations. The 23 Y-STR loci were discovered to be highly polymorphic and suitable for forensic analysis in the Chuxiong Lisu population.
Supplemental Material
Download Zip (476.7 KB)Acknowledgments
We would like to thank the donors who contributed samples for this study.
Disclosure statement
The authors declare that they have no conflicts of interest.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Additional information
Funding
References
- Adnan A, Ralf A, Rakha A, Kousouri N, Kayser M. 2016. Improving empirical evidence on differentiating closely related men with RM Y-STRs: a comprehensive pedigree study from Pakistan. Forensic Sci Int Genet. 25:45–51. doi: 10.1016/j.fsigen.2016.07.005.
- Ballantyne KN, Goedbloed M, Fang R, Schaap O, Lao O, Wollstein A, Choi Y, Van Duijn K, Vermeulen M, Brauer S, et al. 2010. Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications. Am J Hum Genet. 87(3):341–353. doi: 10.1016/j.ajhg.2010.08.006.
- Bar W, Brinkmann B, Budowle B, Carracedo A, Gill P, Lincoln P, Mayr W, Olaisen B. 1997. DNA recommendations. Further report of the DNA commission of the ISFH regarding the use of short tandem repeat systems. International society for forensic haemogenetics. Int J Legal Med. 110(4):175–176. doi: 10.1007/s004140050061.
- Cang M. 1997. On the migration culture of the ethnic groups in Yunnan. Kunming: The Nationalities Publishing House of Yunnan.
- Clegg MT. 1987. Molecular evolution: molecular evolutionary genetics. Science. 235(4788):599. doi: 10.1126/science.235.4788.599.
- Dekairelle AF, Hoste B. 2001. Application of a Y-STR-pentaplex PCR (DYS19, DYS389I and II, DYS390 and DYS393) to sexual assault cases. Forensic Sci Int. 118(2–3):122–125. doi: 10.1016/s0379-0738(00)00481-3.
- Foster EA, Jobling MA, Taylor PG, Donnelly P, De Knijff P, Mieremet R, Zerjal T, Tyler-Smith C. 1998. Jefferson fathered slave’s last child. Nature. 396(6706):27–28. doi: 10.1038/23835.
- Huang Y, Zhang Y, Wang W, Zhao K, Li G, Xu B. 2020. Genetic variation of 15 STR loci in Lisu ethnic minority in southwestern China. Int J Legal Med. 134(2):509–510. doi: 10.1007/s00414-019-02013-4.
- Jobling MA, Tyler-Smith C. 1995. Fathers and sons: the Y chromosome and human evolution. Trends Genet. 11(11):449–456. doi: 10.1016/s0168-9525(00)89144-1.
- Lewis P. 2009. Ethnologue, languages of the world. 16th ed. Dallas: SIL International.
- Lincoln PJ. 1997. DNA recommendations–further report of the DNA commission of the ISFH regarding the use of short tandem repeat systems. Forensic Sci Int. 87(3):181–184.
- Liu GLM, Yu F. 1990. Studies on the physical characters of Lishu nationalty in Yunnan. Acta Anthro Sin. 11(2):8.
- Liu JKW. 2007. Brief on the origin, deposits and changes of the three great clans Di-Qiang, BaiYue and BaiPu. J Yun Agricult Univ (Soc Sci). 1(2):76–78.
- Phillips C. 2015. Forensic genetic analysis of bio-geographical ancestry. Forensic Sci Int Genet. 18(:49–65. doi: 10.1016/j.fsigen.2015.05.012.
- Purps J, Geppert M, Nagy M, Roewer L. 2015. Validation of a combined autosomal/Y-chromosomal STR approach for analyzing typical biological stains in sexual-assault cases. Forensic Sci Int Genet. 19:238–242. doi: 10.1016/j.fsigen.2015.08.002.
- Sun H, Zhou C, Huang X, Liu S, Lin K, Yu L, Huang K, Chu J, Yang Z. 2013. Correlation between the linguistic affinity and genetic diversity of Chinese ethnic groups. J Hum Genet. 58(10):686–693. doi: 10.1038/jhg.2013.79.
- Underhill PA, Kivisild T. 2007. Use of y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu Rev Genet. 41(:539–564. doi: 10.1146/annurev.genet.41.110306.130407.
- Walsh PS, Metzger DA, Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 10(4):506–513.
- Wang Wg DL. 2011. The fusion and disintegration of the ancient di-qiang ethnic group in Southwest China. J Yun National Univ (Soc Sci). 28(3).
- Yin C, He Z, Wang Y, He X, Zhang X, Xia M, Zhai D, Chang K, Chen X, Chen X, et al. 2022. Improving the regional Y-STR haplotype resolution utilizing haplogroup-determining Y-SNPs and the application of machine learning in Y-SNP haplogroup prediction in a forensic Y-STR database: a pilot study on male Chinese Yunnan Zhaoyang Han population. Forensic Sci Int Genet. 57:102659. doi: 10.1016/j.fsigen.2021.102659.
- Zhang X, Zheng H, Liu C, Guo J, Wu J, Hu L, Nie S. 2019. Forensic features for Yunnan Lisu ethnic minority and phylogenetic structure exploration among 26 Chinese populations. Int J Legal Med. 133(1):103–104. doi: 10.1007/s00414-018-1930-5.