?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Background
Inbreeding, arising from consanguinity between related parents, has been observed to impact the health of individuals, typically attributed to biological factors. Nevertheless, these effects may be influenced by the social and environmental conditions. The prevalence of consanguineous marriages increased in certain parts of Sweden after it became legal in 1844, which offers a unique opportunity to study and understand the effects of inbreeding on health.
Aim
The objective of this study is to explore the potential impact of inbreeding on the longevity, fertility, and impairments of individuals born in the Skellefteå region, Sweden, between 1890 and 1905, with a follow-up period extending until 1950.
Subjects and methods
The level of inbreeding is calculated using micro-level parish register data and related to longevity, fertility, and impairments using regression analysis.
Results
Inbreeding is shown to be associated with longevity, fertility, and impairments. It seems to affect the risk of stillbirth and impairments and male longevity and fertility.
Conclusion
Inbreeding seems to have had a detrimental effect on some health outcomes in this historical population under study.
1. Introduction
Inbreeding is the process of reproducing between individuals who are related by blood, implying they share a recent common ancestor. This tends to occur more frequently within populations that are, or have historically been, small in size. Inbreeding can lead to several genetic and biological consequences, including what is known as “inbreeding depression” (Charlesworth and Willis Citation2009). Inbreeding increases the frequency of homozygous alleles, where identical alleles are present at a specific gene locus. This heightens the probability that individuals will inherit two copies of a recessive deleterious allele from their common ancestor, increasing the risk of genetic disorders or congenital diseases. Inbreeding depression can manifest as reduced overall health, resulting in decreased reproductive success, lower survival rates, and poorer health in the offspring. In outbred populations, the presence of dominant alleles often conceals these recessive alleles.
A vast literature exists describing the effects of inbreeding. Rudan et al. (Citation2003) demonstrated that inbreeding is a predictor of conditions like coronary heart disease, stroke, cancer, unipolar/bipolar depression, asthma, gout, and peptic ulcer. They suggest that a large part of these diseases resulting in lifelong impairments can be caused by inbreeding in societies where consanguineous marriages are common. The mechanism is suspected to operate through the homozygosity of deleterious recessive alleles.
However, inbreeding cannot be seen as purely a biological phenomenon. Richardson and Smiseth (Citation2023) argue that social interactions, such as cooperation, competition, and communication by either exacerbating or buffering against inbreeding depression. At a societal level avoidance mechanisms, such as cultural and religious taboos on incest, and laws explicitly forbidding certain family formation also affect the situation for the families. This means that what is observed in empirical studies is shaped not only by genetics and biology but also by social, cultural, and environmental circumstances. This highlights the importance of studying the effect of inbreeding in varying settings to gain a better understanding of the mechanisms of how it affects different outcomes.
Studying the effect of inbreeding can be challenging in practice due to the complexity of constructing adequately large pedigrees, primarily stemming from the limited historical data depth available in most registers. In addition to that, the registers must allow a long follow-up period to possibly identify and assess the effects on the individuals. These obstacles are overcome in this study using extensive Swedish registers going back to the late seventeenth century.
The aim of the paper is to investigate whether the inbreeding level is associated with health outcomes for individuals in a regional population in Sweden born 1890–1905 and followed for 45 years, at the longest until 1950. To achieve this, data from parish records from the Skellefteå region, covering individuals from 1680 to 1950, a time depth of 270 years, are used to generate pedigrees and calculate the level of inbreeding. In the study area, the Skellefteå region in Sweden, the level of consanguineous marriages was relatively high in the nineteenth century, especially after cousin marriages became legal in 1844 (Inger Citation1980; Widerberg Citation1980).
To get a comprehensive picture of how inbreeding affected human life during this period the effect on longevity, fertility, and impairments are investigated.
The same region has been used before to study inbreeding (Bittles and Egerbladh Citation2005; Egerbladh and Bittles Citation2008). Even though Bittles and Egerbladh (Citation2005) discuss the potential effect of inbreeding on genetic disorders in the Skellefteå region, they do not explicitly link inbreeding with impairments as we do in this study. Egerbladh and Bittles (Citation2008) study the effect of inbreeding 1720–1899 and conclude that it has an effect on stillbirths and infant and early childhood mortality but no effect on fertility. In the present study, we analyse the cohort born 1890–1905 with a longer follow-up period, stretching well into the twentieth century, accounting for a wider set of outcome variables. In addition, we consider the pedigrees from the start of the recording of sources which has been facilitated by the linking of the genealogical information in the different parishes in the region. This enables us to consider even small and accumulated inbreeding in the population.
Our analysis contributes to research analysing the effects of inbreeding on fertility and longevity by basing the results on a past Swedish population followed over centuries and by considering the inbreeding effects on health outcomes represented by impairments. It is important to study the effects of inbreeding in various socio-economic settings because the surrounding circumstances can impact its influence. Our results are relevant for history, demography, and medicine in trying to understand the consequences of inbreeding by identifying and assessing how it affects different health outcomes.
2. Materials and methods
2.1. Materials
The data for this study are sourced from parish registers that are maintained by the Demographic Data Base (DDB) at Umeå University (Westberg et al. Citation2016). These registers contain digitised and linked parish records, originating from the actual registers that document parishioners’ vital events, such as birth, baptism, marriage, migration, death, burial, and catechetical examination records. Our dataset encompasses longitudinal and multi-generational data, recorded for a total of 227,215 parishioners who resided in the Skellefteå region from 1680 to 1950. These registers are not only linked at the individual level but also interconnected with records of preceding generations across parishes. This linkage provides exceptionally comprehensive demographic data into the lives of each parishioner, including their family background. The wealth of data allows us to trace family histories back for nearly three centuries, to construct pedigrees for the entire population, which, in turn, facilitates the calculation of inbreeding levels. With the ability to follow individuals for a significant portion of their lives, we can detect and assess the effects of inbreeding.
The birth and death registers provide valuable information on fertility and mortality within the population. For identifying impairments in individuals over their lifetime, the catechetical registers are crucial. These registers were collected on an annual basis, as ministers were obligated to maintain records of parishioners’ knowledge of the catechism and their reading abilities. This obligation was initially established in the Church law of 1686 (Nilsson Citation1994). In addition to educational and religious information, these catechetical examination records include marks of impairments (lytesmarkeringar) following guidelines from Statistics Sweden and medical expertise at the time. These marks document physical, sensory, and mental deviations in functionality among the parishioners and are essential for identifying impaired individuals and distinguishing them from non-impaired cases (Rogers and Nelson Citation2003; Haage Citation2017; Wisselgren and Vikström Citation2023). Thus, we could identify and code this impairment information into a binary variable, here focusing on impairment reported in ages up to 45. It should be noted that the quality of the data does not allow us to distinguish between impairments due to genetic causes and other causes which adds a disturbance but should not systematically change the results of the analysis.
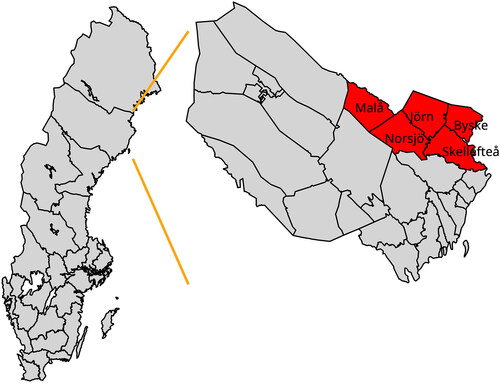
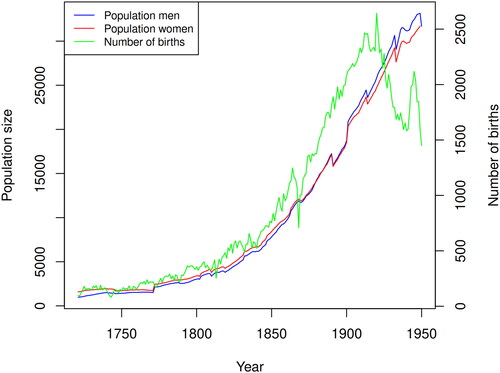
The region under study consists of the Skellefteå town and rural parish and five nearby parishes in northern Sweden as depicted in . The socio-economic structure was mainly based on agricultural production with increasing urbanisation of the town of Skellefteå from the early twentieth century. The population in northern Sweden has a history of admixture of Swedish, Sami, and Finnish peoples and has experienced a dramatic population growth based on a relatively small founding population (Einarsdottir et al. Citation2007). illustrates the population trends from 1721 to 1950. The sharp discontinuities in the population size are primarily caused by changes in administrative boundaries among the parishes while the sharp drop in the number of births in the 1860s was due to a famine. Key drivers of demographic change were wars and famines, leading to population bottlenecks and reduced genetic diversity.
The relatively small founding population is evident in , which displays the number of births in the region from 1700 to 1950. The birth rate increased from 83 per year in 1700 to 1449 per year in 1950, reaching its peak in 1920 when 2643 births were recorded. The small founding population results in a “pedigree collapse” or “founder effect” according to which the number of distinct ancestors in the family tree of individuals is smaller than it could be due to the recurrence of the same ancestors in multiple places within the pedigree. This effect is further exacerbated by the tendency to marry within the same area, such as a river valley, which leads to the creation of sub-isolates. These sub-isolates have been confirmed through DNA tests (Einarsdottir et al. Citation2007).
Figure 2. Population development in the Skellefteå region. Total number of parishioners by sex and children born ca. 1700–1950.
Data source: Digitised parish register, Demographic Data Base, Umeå University.
Our dataset comprises observations of individuals who resided in the Skellefteå region at some point during the time when parish records were maintained. To calculate the inbreeding factor we use pedigrees as far back as the registers allow. A descriptive summary of the data can be found in in Appendix A.
The primary focus is on individuals noted as present in the Skellefteå region and born between 1890 and 1905 and have complete three generation pedigrees. This amounts to 8191 live births, and 198 still-born and will serve as the study population for the longevity models where the individuals are followed until death or censoring. Noteworthy is that only about 2215/8192 = 27% has an inbreeding factor of zero and that almost half of the study population (4058/8192) has a inbreeding factor above zero but below 1/256. The requirement of three generations of full pedigree leads to a population with deep roots in the region with many possibilities of parents to be distantly related.
Out of these individuals, 3060 can be followed from birth to the age of 45 without any absences longer than a year and they will serve as the study population for analysing aspects related to fertility and impairment. This will encompass the main fertile period and allow impairment to have time to manifest itself in individuals and at the same time not include impairments that come with old age.
From the variables available in the data retrieval, it is possible to derive several key variables for our analysis, such as age of death, number of children, whether there is an impairment note, and also to calculate individual inbreeding coefficients and the inbreeding coefficient of the children to a father.
2.2. Methods
The investigation of the effects of inbreeding was conducted using the so called inbreeding coefficient as the key variable under study. The inbreeding coefficient, denoted as F, is a measure that quantifies the level of inbreeding in an individual with a larger value indicating a higher extent of inbreeding (Ballou Citation1983). F is defined as the probability that two alleles at a specific gene locus are identical by descent, meaning they come from a common ancestor. If this allele defines a recessive genetic disease it can be interpreted as the probability that it will manifest itself in the offspring. This coefficient provides a numerical representation of the extent of inbreeding and has proven useful for differentiating the impact of inbreeding on various aspects of the population. For example, parents that are full siblings get children that have F = 1/4, first cousins F = 1/16, second cousins F = 1/64, and third cousins F = 1/256. It should be noted that the calculation of F is based on available data from the parish registers, potentially resulting in an underestimate of the true inbreeding levels when the inbreeding event predates the available data.
In clinical genetics, consanguineous marriage is typically defined as couples who are related as second cousins or closer (Modell and Darr Citation2002). However, in population genetics, any level of shared ancestry in a couple is often considered a consanguineous marriage. For this study, we were interested in even minor inbreeding resulting from consanguineous relationships, making the broader definition more applicable. In this context, inbreeding can be considered the reproductive consequence of a consanguineous marriage.
The inbreeding coefficient, F, was the primary study variable used to investigate its effect on health outcomes related to longevity, fertility, and impairments. F was categorised into five inbreeding groups, with the null group serving as the reference level in the regression models, as described in . The frequency in each group is also shown, indicating a sufficient number in each group. In most models, the individuals’ own inbreeding coefficient was used. However, when the effect of couples being related was studied, the mean inbreeding coefficient of the children of the father was used along with the number of children of that father. The reason for this was to handle remarriages leading to different inbreeding coefficients of the children. The cases of remarriages are relatively small and will have low impact on the analysis.
Table 1. Inbreeding groups.
The analysis is carried out with different regression models as outlined in . The linear part in the regression models is defined in the same way in all the models and shown in Formula 1:
(1)
(1)
where:
Table 2. Models estimated by type of outcome under investigation.
The birth year variable was used to adjust for potential fluctuations due to other factors associated with time that could otherwise affect the estimation of the parameters related to inbreeding. The variable indicating if the father of the individual was a farmer can potentially capture some of the social differences in the population. All analyses were made in R (R Core Team Citation2022), where the calculation of the inbreeding factor was made using the calcInbreeding function in the pedigree package (Coster Citation2022).
The remaining part of the subsection describes the models used to investigate longevity, fertility, and impairments with a summary in .
Longevity was measured as the total number of days lived. The effect of inbreeding on longevity was investigated using Cox regressions where separate models were estimated for both sexes and three distinct age intervals: 0–28 days, 29–365 days, and periods exceeding one year, denoted 1a–c for men and 2a–c for women. This stratification was based on the assumption that the effect of inbreeding might vary across these age categories and between sexes. The age groups follow the intervals used by, for example, the World Health Organisation and can be motivated by the change in causes of death as individuals age. The linear part of the model is shown in Formula 1. In a Cox regression model, the coefficients quantify the relationship between explanatory variables and the hazard rate. A positive coefficient suggests an increased risk of an event, here death, while a negative coefficient indicates a reduced risk associated with the variable it represents. We analysed stillbirths in a similar way but using logistic regression and the model in Formula 1 (Model 3). The child’s own inbreeding group was the study variable. The outcome variable indicates if the birth was a stillbirth and higher parameter estimates indicate an increased risk.
Fertility was measured by the number of biological children a person has. Models 4a and 4b are concerned with the effect on fertility of the inbreeding factor the mother or father have themselves. The modelling approach was zero inflated models with a logistic regression part to model the excess zeros and a Poisson model for the count part similar to the model proposed in Álvarez et al. (Citation2015). This approach adjusts for excess zeros in the data increasing the validity of the following Poisson model (Models 4a–b). The logistic part models the probability of not having a child, where a positive parameter estimate indicates a higher probability of not having a child. In the Poisson model, a positive parameter estimate indicates a tendency to have more children. All models used in the fertility analysis are defined by Formula 1.
The model concerning the fertility of couples (Model 4c) used the mean of children inbreeding to a specific father as measure of parents’ relatedness. As outcome variable the number of children above one of that father is used, thus requiring at least one child to have been born. This approximates the parents’ genetical relatedness and number of children. A Poisson model is used for Model 4c as it does not have the problem of excess zeros. This model requires data on both parents’ pedigrees to be sufficient making the total number of observations slightly lower. The inbreeding group was calculated as the mean inbreeding factor of children to a father and the number of children is the father’s number of children.
To assess the impact of inbreeding on impairments, we relied on impairment notes in the registers as the outcome using logistic regression and the linear as in Formula 1 (Model 5). Common notes found in the parish registers include sensory impairments like deaf, mute, and blind; physical impairments like crippled; and mental disorders like insane or idiot. The outcome variable was an impairment note of any kind, recorded as a binary variable, and analysed with logistic regression using the model in Formula 1. Positive parameter estimates indicate higher propensity to have an impairment. A minimum follow-up duration of 45 years was required for this analysis due to the potential for late-onset conditions associated with inbreeding (Rudan et al. Citation2003).
3. Results
This section presents the results of the different model estimates. The major findings are illustrated by graphs, while complete estimates are detailed in the tables in Appendix A.
3.1. The effect of inbreeding on longevity
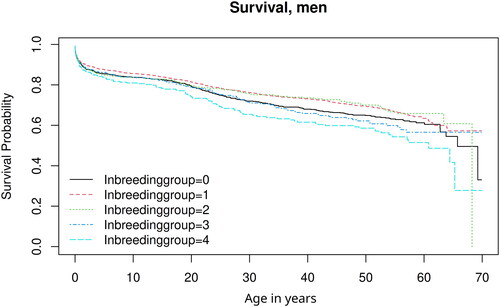
reports the result for the longevity/mortality estimates for men. The inbreeding group variable for Models 1a and 1b is not significant according to the results of the Cox regression. In Model 1c the higher inbreeding group 4 has significantly higher mortality indicating a detrimental effect of inbreeding for men above the age of one. In this model the Likelihood ratio tests (LR-test) are significant, testing the complete inbreeding variable and not just the levels, indicating an effect of inbreeding. This result is visible in the Kaplan–Meier curves of showing a lower survival for inbreeding group 4. In this model, the dummy variable denoting if the father is a farmer is also significant indicating a protective effect. This suggests that the relatively wealthy socio-economic situation of the farmer families was beneficial for survival. The results for women shown in were not significant for inbreeding according to the Likelihood ratio tests (LR-test) testing the complete inbreeding variable and not just the levels.
Figure 3. Kaplan–Meier survival curves showing men’s longevity by own inbreeding group.
Source: Digitised parish register, Demographic Data Base, Umeå University.
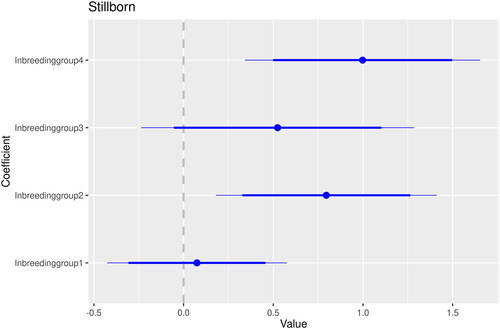
shows that consanguineous parents had an increased risk of getting stillborn children with the most inbred at highest risk. The parameter estimates for inbreeding group are illustrated in . The parameter estimates can be translated to estimated probabilities suggesting that a child born year 1900 with a farming father has a risk of being stillborn of 1.9% in inbreeding group 0, 2.1% in group 1, 4.2% in group 2, 3.2% in group 3, and 5.1% in group 4.
3.2. The effect of inbreeding on fertility
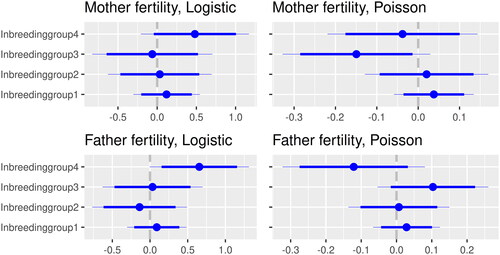
The estimates of the logistic models are presented in while shows the results of the Poisson part. The estimates of the effect of inbreeding are summarised in . There was an evident tendency for both men/fathers and women/mothers in the highest inbreeding group to have zero children as can be seen in Models 4a and 4b although it is only significant at the 5% level for men. The Poisson part for the number of children is not significant for these models. Moreover, the inbreeding group is not significant for the couples relatedness model either (Model 4c).
3.3. The effect of inbreeding on impairments
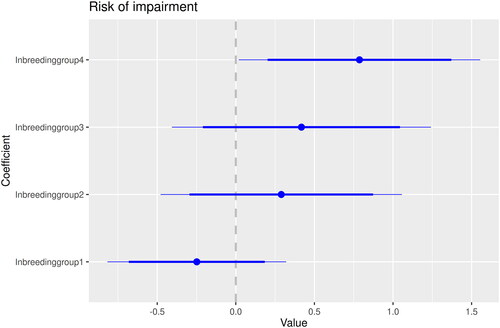
The results on impairments are significant for inbreeding group according to the LR-test. Thus, there was an increasing risk for impairment with the level of inbreeding (). The parameter estimates are visualised in . These estimates can be translated to estimated probabilities, according to which a child born year 1900 with a farming father has a risk of being impaired of 4.6% in inbreeding group 0, 3.6% in group 1, 6.0% in group 2, 6.8% in group 3, and 9.6% in group 4.
4. Discussion
In this study, we investigate three health aspects of human life that can be affected by inbreeding. We find that high inbreeding affects male longevity more than women’s, with a significantly higher mortality above one year of age for those with higher inbreeding levels. The results for women and ages below one year among both sexes were not significant. This observation aligns with Helgason et al. (Citation2008), who showed that for the Icelandic population, the life expectancy of children of couples who were related as second cousins or closer was shorter than for couples with less inbreeding. In our study, the inbreeding effect is more pronounced for the risk of stillbirths, where the estimated risk increases from 1.9% in the group with no inbreeding to 5.1% in the highest group. This is in line with previous research by Stoltenberg et al. (Citation1999). One explanation for the seemingly low effect on neonatal and infant mortality might be the high overall mortality rates in these age groups, making the specific effect of inbreeding harder to detect.
Several scholars have examined the effect of inbreeding on human fertility with mixed results. Studies that found negative effects include Robert et al. (Citation2009) and Álvarez et al. (Citation2015). Postma et al. (Citation2010) note that related couples did not have fewer children themselves, while their inbred daughters did. The effect of inbreeding on fertility found in this study was that men with the highest levels of inbreeding were more likely to not have children. The number of children, given that the individual had at least one child, was not significant, as evidenced by the Poisson models for both men and women. This is consistent with the findings of Robert et al. (Citation2009), who reported a negative relationship between fathers’ inbreeding and fertility. The effect of parents being related was not significant. Thus, our findings do not support the results in the study from Iceland by Helgason et al. (Citation2008), where couples related but less so than fourth cousins had more children compared to those who were not inbred.
One factor that might affect our fertility analysis is reproductive compensation. This is a phenomenon where individuals increase their reproductive efforts in response to a decline in reproductive success. Signs of reproductive compensation were found in the inbreeding study by Ober et al. (Citation1999). This might mask the negative effects of inbreeding also concerning our fertility findings.
We find in this study that inbreeding has a rather strong effect on the risk of acquiring an impairment. For example, Rudan et al. (Citation2003) demonstrated that inbreeding is a predictor of conditions resulting in lifelong impairments. Our study indicates that this association exists.
Our study suggests that the effect of having a father who was a farmer was significant in one model, for male longevity above one year of age. This indicates that socioeconomic status might moderate the effect of inbreeding, but this effect was not consistent across outcomes investigated here. Having a farming father was the dominant socioeconomic group, comprising 6889 out of 8191 individuals, which is 84%. The study design, requiring a full three-generation pedigree, contributed to this, as the farming community is more geographically stable. The mean inbreeding coefficient for children of non-farmers was 0.0022, and for farmers’ children, it was 0.0049, more than double as high. Having a farming father was thus associated with inbreeding and was itself a factor that affected the outcome variables, making it a potentially confounding factor and an important variable to consider. Future studies can make a more thorough study of the effect of inbreeding on different socioeconomic strata in the population. We find that this would require more data than is currently available, as other socioeconomic groups are too small to be used to investigate the effect of inbreeding. In a more modern context, Johnson et al. (Citation2018) found relationships between estimated autozygosity and complex traits using the UK Biobank data. Some relationships are attenuated after controlling for background sociodemographic characteristics. This indicates that such characteristics are important to control for if possible.
Studying the same Skellefteå region as in our study, Egerbladh and Bittles (Citation2011) encountered a non-random pattern in first cousin marriages. They found significantly more consanguineous marriages among land-owning families, especially involving first-born sons, within specific pedigrees, and in several more remote inland communities. They also speculated that consanguineous marriages are more likely to occur due to factors other than just being a farmer, such as physical or mental impairments among males and, among females, the previous birth of a child out of wedlock. This proposition suggests that inbreeding can result from health and social problems, not only be the cause of them. This potentially bidirectional dependence is challenging to control for with the data at hand in the present study but calls for caution in interpreting the results.
In addition, there are, of course, a wealth of variables not controlled for in this study that affect our outcome variables: longevity, fertility, and impairment. These can, for example, include the individual’s health status or environmental factors that affect the outcome variables through more or less complex mechanisms. If these are also associated with the level of inbreeding in a systematic way, they can distort the results. In our study, we are unable to include such variables, but it should be remembered that there might exist variables that can modify the observed effects in the models.
Another potential problem could be that the outcome variables might be dependent on each other. For example, deaths before the follow-up period of 45 years disqualify individuals from the fertility and impairment analysis, resulting in a potentially healthier subset for our analyses. This could lead to an underestimation of the detrimental effects of inbreeding.
Hence, our approach taken to measure inbreeding by the inbreeding coefficient calculated from an observed pedigree created from parish registers has some limitations. The pedigree can be incorrect due to errors, such as recording the wrong parent in the original file or errors in digitisation. The variables measured concerning fertility and mortality are likely to be of good quality as they are easy to measure and record. However, the impairment variable is less consistent and not as systematically recorded, which might lead to under-reporting of such cases. Additionally, in the sources, we have not been able to distinguish between impairments caused by genetic effects and those caused by other factors, such as accidents or diseases.
Moreover, the time depth is limited by the start of the registers, likely leading to an underestimation of inbreeding in the population. This limitation can contribute to the small differences between those that we have calculated to be inbred and the non-inbred population. The population that we regard as non-inbred might, therefore, in reality, be inbred to some extent. An alternative approach to overcome such problems would be to use DNA tests from the population, an approach taken in studies, such as Karafet et al. (Citation2015). However, this approach has the drawback of normally resulting in a much smaller data set than the full population registers used in the present study.
One sign of unrecorded inbreeding in the Skellefteå region can be that some genetic diseases, both autosomal recessive and autosomal dominant, have a relatively high frequency. Nordström (Citation1991) provides an overview of these genetic diseases, one of which is well-known and referred to as “Skelleftesjukan” or “The Skellefteå sickness,” formally called Hereditary Transthyretin Amyloidosis (Norgren Citation2014). It’s origin was traced back to six ancestral couples born between 1599 and 1736. Affected cases are predominantly concentrated in the Skellefteå area, highlighting the limited influx of people from outside the region. This can point to unrecorded and underestimated inbreeding.
Similar effects have been found in other studies in that base their results on a population with limited genetic exchange with other populations. This is the case for the Icelandic population used by Helgason et al. (Citation2008). Another isolated island population that has been studied, for example, by Macgregor et al. (Citation2010), is that of Norfolk Island, known for the mutiny on the Bounty in 1789. In their study, inbreeding is related to various cardiovascular disease traits. O’Brien et al. (Citation1988) conducted a study in yet another island context, focusing on rare genetic diseases in an isolated parish in the Finnish archipelago region of Åland, using old parish registers to identify the founders. The prevalence of von Willebrand disease and tapetoretinal disease is high within this island community. They conclude that this is caused by a high frequency of the causative genes in the founding population or by repeated reintroduction through migration, rather than by a small number of individuals in the founding population. Dorsten et al. (Citation1999) found a net positive effect on neonatal and post-neonatal deaths among inbred children in the Amish population. They suggest social, demographic, and population-based socio-cultural explanations for this effect.
5. Conclusions
This research delves into how inbreeding affects the health of individuals born in Skellefteå, Sweden, from 1890 to 1905, with a follow-up period until 1950. By analysing detailed parish register data at the individual level, digitised from the original written sources, the study examines how inbreeding relates to longevity, fertility, and impairments using various regression analysis techniques. Pedigrees can be constructed stretching hundreds of years back in time, which, in combination with a relatively high proportion of marriages between relatives in the nineteenth century, allows us to study the health outcomes of inbreeding. Results indicate connections between inbreeding and these health metrics within this historical context, suggesting a noticeable but modest impact, potentially influenced by socio-economic variables. This paper enriches existing literature by offering insights into the effects of inbreeding from various angles in this specific historical context. Our study is the first analysis to look for evidence on whether and how inbreeding historically affects impairments using parish registers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Álvarez G, Ceballos FC, Berra TM. 2015. Darwin was right: inbreeding depression on male fertility in the Darwin family. Biol J Linn Soc Lond. 114(2):1–9. doi: 10.1111/bij.12433.
- Ballou JD. 1983. Calculating inbreeding coefficients from pedigrees. Genet Conserv. 509:520.
- Bittles A, Egerbladh I. 2005. The influence of past endogamy and consanguinity on genetic disorders in northern Sweden. Ann Hum Genet. 69(Pt 5):549–558. doi: 10.1046/j.1529-8817.2005.00179.x.
- Charlesworth D, Willis JH. 2009. The genetics of inbreeding depression. Nat Rev Genet. 10(11):783–796. doi: 10.1038/nrg2664.
- Coster A. 2022. Pedigree: pedigree functions. R package version 1.4.2. https://CRAN.R-project.org/package=pedigree.
- Dorsten LE, Hotchkiss L, King TM. 1999. The effect of inbreeding on early childhood mortality: twelve generations of an Amish settlement. Demography. 36(2):263–271. doi: 10.2307/2648113.
- Egerbladh I, Bittles A. 2008. The influence of consanguineous marriage on reproductive behavior and early mortality in northern coastal Sweden, 1780–1899. In: Kinship and demographic behavior in the past. International studies in population. Dordrecht: Springer; p. 205–224.
- Egerbladh I, Bittles A. 2011. Socioeconomic, demographic and legal influences on consanguinity and kinship in northern coastal Sweden 1780–1899. J Biosoc Sci. 43(4):413–435. doi: 10.1017/S0021932011000125.
- Einarsdottir E, Egerbladh I, Beckman L, Holmberg D, Escher SA. 2007. The genetic population structure of northern Sweden and its implications for mapping genetic diseases. Hereditas. 144(5):171–180. doi: 10.1111/j.2007.0018-0661.02007.x.
- Haage H. 2017. Disability in individual life and past society: life-course perspectives of people with disabilities in the Sundsvall region of Sweden in the nineteenth century [PhD dissertation]. Umeå: Umeå University.
- Helgason A, Pálsson S, Gudbjartsson DF, Kristjánsson T, Stefánsson K. 2008. An association between the kinship and fertility of human couples. Science. 319(5864):813–816. doi: 10.1126/science.1150232.
- Inger G. 1980. Svensk rättshistoria. Stockholm: Liber.
- Johnson EC, Evans LM, Keller MC. 2018. Relationships between estimated autozygosity and complex traits in the UK Biobank. PLOS Genet. 14(7):e1007556. doi: 10.1371/journal.pgen.1007556.
- Karafet TM, Bulayeva KB, Bulayev OA, Gurgenova F, Omarova J, Yepiskoposyan L, Savina OV, Veeramah KR, Hammer MF. 2015. Extensive genome-wide autozygosity in the population isolates of Daghestan. Eur J Hum Genet. 23(10):1405–1412. doi: 10.1038/ejhg.2014.299.
- Macgregor S, Bellis C, Lea RA, Cox H, Dyer T, Blangero J, Visscher PM, Griffiths LR. 2010. Legacy of mutiny on the bounty: founder effect and admixture on Norfolk Island. Eur J Hum Genet. 18(1):67–72. doi: 10.1038/ejhg.2009.111.
- Modell B, Darr A. 2002. Genetic counselling and customary consanguineous marriage. Nat Rev Genet. 3(3):225–229. doi: 10.1038/nrg754.
- Nilsson H. 1994. Mot bättre hälsa: dödlighet och hälsoarbete i Linköping 1860–1894 [PhD dissertation]. Linköping: Linköpings Universitet.
- Nordström S. 1991. Genealogi och genetik: beskrivning av sex projektområden där kyrkböcker använts i genetisk forskning. Forskningsarkivet. Umeå: Umeå Universitet.
- Norgren N. 2014. Hereditary transthyretin amyloidosis (ATTR V30M): from genes to genealogy [PhD dissertation]. Umeå: Umeå Universitet.
- O’Brien E, Jorde LB, Rönnlöf B, Fellman JO, Eriksson AW. 1988. Founder effect and genetic disease in Sottunga, Finland. Am J Phys Anthropol. 77(3):335–346. doi: 10.1002/ajpa.1330770306.
- Ober C, Hyslop T, Hauck WW. 1999. Inbreeding effects on fertility in humans: evidence for reproductive compensation. Am J Hum Genet. 64(1):225–231. doi: 10.1086/302198.
- Postma E, Martini L, Martini P. 2010. Inbred women in a small and isolated Swiss village have fewer children. J Evol Biol. 23(7):1468–1474. doi: 10.1111/j.1420-9101.2010.02013.x.
- R Core Team. 2022. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. https://www.R-project.org/.
- Richardson J, Smiseth PT. 2023. A behavioral ecology perspective on inbreeding and inbreeding depression. Adv Study Behav. 55:37–54.
- Robert A, Toupance B, Tremblay M, Heyer E. 2009. Impact of inbreeding on fertility in a pre-industrial population. Eur J Hum Genet. 17(5):673–681. doi: 10.1038/ejhg.2008.237.
- Rogers J, Nelson MC. 2003. “Lapps, Finns, gypsies, jews, and idiots”? Modernity and the use of statistical categories in Sweden. Ann Démogr Hist. 105(1):61–79. doi: 10.3917/adh.105.79.
- Rudan I, Rudan D, Campbell H, Carothers A, Wright A, Smolej-Narancic N, Janicijevic B, Jin L, Chakraborty R, Deka R, et al. 2003. Inbreeding and risk of late onset complex disease. J Med Genet. 40(12):925–932. doi: 10.1136/jmg.40.12.925.
- Stoltenberg C, Magnus P, Skrondal A, Lie RT. 1999. Consanguinity and recurrence risk of stillbirth and infant death. Am J Public Health. 89(4):517–523. doi: 10.2105/ajph.89.4.517.
- Westberg A, Engberg E, Edvinsson S. 2016. A unique source for innovative longitudinal research: the POPLINK database. Hist Life Course Stud. 3:20–31. doi: 10.51964/hlcs9351.
- Widerberg K. 1980. Kvinnor, klasser och lagar 1750–1980. Stockholm: LiberFörlag.
- Wisselgren MJ, Vikström L. 2023. Behind the numbers: authorities’ approach to measuring disability in Swedish populations from 1860 to 1930. Hist Methods. 56(2):63–76. doi: 10.1080/01615440.2023.2186998.
Appendix A
Table A1. Summary of the data, frequencies.
Table A2. Model 1a–c, Cox regression, longevity male.
Table A3. Model 2a–c, Cox regression, longevity female.
Table A4. Model 3, logistic regression, stillbirths.
Table A5. Model 4a and 4b, logistic part of zero inflated model, fertility.
Table A6. Model 4a, 4b, and 4c, Poisson regression, fertility.
Table A7. Model 5, logistic regression, impairment/disability.