Abstract
Forty-two isolates of Verticillium dahliae were recovered from stem and root samples of olive trees showing symptoms of verticillium wilt in various olive-growing regions in Tunisia. Each isolate was identified based on microscopic observations of morphological and cultural characteristics, pathogenicity tests, as well as PCR amplification using Vd1/Vd2 primers. Genetic diversity among the isolates was investigated using random amplified microsatellites (RAMS) and PCR-RFLP of intergenic spacer region (IGS) of ribosomal DNA (rDNA). A single fragment of approximately 1.7–2.1 kb was amplified from all isolates by PCR using primers CNL12 and CNS1. Digestion of the amplified IGS region with restriction enzyme RsaI produced similar banding patterns (1200 and 800 bp) for 40 isolates while individual and distinctive banding patterns (1100, 850 and 150 bp) were observed for two isolates. Using RAMS primers, nine, eight and four bands were produced when using primers CGA (2200, 1400, 1200, 1100, 1000, 650, 550, 500 and 350 bp), CCA (2000, 1200, 950, 850, 800, 550, 500 and 400 bp) and GT (2500, 2400, 700 and 500), respectively. A total of 21 polymorphic markers were scored when data from the RAMS experiments were combined. Overall, the results of this study revealed associations between the genetic diversity of the isolates and their pathogenicity phenotype, but not between genetic diversity and the geographic origin, suggesting that they are randomly spread across Tunisia.
Résumé
Dans différentes régions de la Tunisie, 42 isolats de Verticillium dahliae ont été récupérés d’échantillons de tiges et de racines d’oliviers montrant des symptômes de flétrissure verticillienne. Chaque isolat a été identifié en fonction d’observations au microscope des caractères morphologiques et culturaux, de tests de pathogénicité ainsi que de l’amplification par PCR avec les amorces Vd1/Vd2. La diversité génétique au sein des les isolats a été étudiée par amplification aléatoire des microsatellites (RAM) et par PCR-RFLP de la région de l’espaceur intergénique (IGS) de l’ADN ribosomique (ADNr). Un seul fragment d’environ 1.7 à 2.1 kb de chaque isolat a été amplifié par PCR avec amorces CNL12 et CNS1. La digestion de l’IGS amplifiée par l’enzyme de restriction Rsal a produit des spectres de bandes similaires (1200 et 800 bp) chez 40 isolats, tandis que des spectres de bandes individuels et particuliers (1100, 850 et 150 bp) ont été observés pour deux isolats. En utilisant les amorces RAM, neuf, huit et quatre bandes ont été produites respectivement avec les amorces CGA (2200, 1400, 1200, 1100, 1000, 650, 550, 500 et 350 bp), CCA (2000, 1200, 950, 850, 800, 550, 500 et 400 bp) et GT (2500, 2400, 700 et 500 bp). En tout, 21 marqueurs polymorphiques ont été détectés quand les données des expériences basées sur la méthode RAM ont été combinées. Dans l’ensemble, les résultats de cette étude ont révélé des liens entre la diversité génétique des isolats et leur phénotype de pathogénicité, mais pas entre la diversité génétique et l’origine géographique, ce qui suggère qu’ils sont répartis aléatoirement sur tout le territoire tunisien.
Introduction
Olive is a very important crop in many Mediterranean countries. In Tunisia, it represents 6.7% of the world olive production (FAO Citation2008), and is constantly at risk of infection by the soilborne pathogen Verticillium dahliae Kleb. This fungus causes a vascular wilt disease which has been extensively studied in many important crops and forest tree species (Bhat & Subbarao Citation1999; Pegg & Brady Citation2002; Barbara & Clewes Citation2003; Robb Citation2007; Klosterman et al. Citation2009; El Hadrami et al. Citation2011). In Tunisia, V. dahliae was first reported on olive in 2006 by Triki et al. (Citation2006), and shared morphological, physiological and virulence features common to Verticillium species affecting other crop plants (Jabnoun-Khiareddine et al. Citation2006). Useful information on the diversity of V. dahliae populations has been provided by vegetative compatibility groupings (VCG), especially when combined with pathogenicity studies (Daayf et al. Citation1995; Dobinson et al. Citation2000; Korolev et al. Citation2008; Alkher et al. Citation2009). However, understanding the complex population structure of this fungus and its ability to infect different crops cannot be achieved using these techniques alone and usually requires further molecular analysis (El-Bebany et al. Citation2010, Citation2011, Citation2013).
Different methods have been used to identify molecular markers that are best associated with genetic variation in V. dahliae. For example, isolates from olive trees have been studied using RAPD analysis (Cherrab et al. Citation2000; Bellahcene et al. Citation2005). While this technique successfully correlated the geographic origin of a collection of olive-infecting isolates in Morocco (Cherrab et al. Citation2000), limited genetic diversity was found among isolates collected across the border in Algeria (Bellahcene et al. Citation2005). More recently, genetic diversity among V. dahliae populations was investigated using AFLP analysis (Collado-Romero et al. Citation2006; Erdogan et al. Citation2013), as well as the relationship of this molecular marker to VCGs in isolates infecting artichoke, cotton and olive. Analysis and comparison of DNA sequences such as the intergenic (IGS) and internal transcribed (ITS) spacer regions of rDNA have been used to differentiate, and in some cases, detect and quantify, Verticillium isolates from diverse host plants and soil samples (Nazar et al. Citation1991; Morton et al. Citation1995; Bilodeau et al. Citation2012). The IGS region of nuclear rRNA repeat units evolve faster and may vary both among and within species (Bainbridge Citation1994). In this context, Pramateftaki et al. (Citation2000) detected great heterogeneity in the IGS region through analysis of the complete ribosomal DNA sequence of Verticillium strains isolated from different hosts and locations. Further phylogenetic analyses of IGS rDNA sequences coupled with virulence data suggested a relationship between phylogenetic grouping and virulence for some strains of V. dahliae (Qin et al. Citation2006). More recently, structural and phylogenetic analyses of the IGS region identified two main lineages in V. dahliae, with clade I including VCGs 1A, 1B, 2A, 4B and 3 and clade II containing VCGs 2B, 4A and 6 (Papaioannou et al. Citation2013). PCR-RFLP of these regions has also been used to differentiate isolates of V. dahliae (Papaioannou et al. Citation2013). The random amplified microsatellites (RAMS), another PCR-based technique first used in plants and animals (Zietkiewicz et al. Citation1994), then in fungi (Hantula et al. Citation1996), combines the simplicity of RAPD with the efficiency of microsatellite analyses, and has the potential to differentiate populations or recently diverged species (Zhou et al. Citation2001; Ratanacherdchai et al. Citation2010). This technique was useful in describing genetic diversity in several groups of fungi such as Claviceps spp. (Tooley et al. Citation2000), Botryosphaeria spp. (Zhou et al. Citation2001), Phaeoisariopsis griseola (Mahuku et al. Citation2002), Fusarium culmorum (Mishra et al. Citation2003), Gremmeniella spp. and Phomopsis spp. (Børja et al. Citation2006) and the oomycetes Phytophthora cactorum (Hantula and Müller Citation1997) and P. citrophthora (Cohen et al. Citation2003), but has not been previously used in Verticillium.
Despite the extent and importance of olive production in Tunisia and the threat from verticillium wilt, local populations of V. dahliae have not been studied from a molecular point of view. Determining the status of such diversity is essential to select suitable resistant olive cultivars for specific growing regions, and to monitor changes in the structure of the pathogen populations that represent a threat to olive cropping in Tunisia and other Mediterranean countries. The objectives of this study were to (i) investigate the genetic diversity of 42 isolates of V. dahliae collected from infected olive trees in the most important olive growing regions in Tunisia; and (ii) test the hypothesis of a correlation between the pathogenicity phenotype, geographic origin and molecular markers. For this purpose, pathogenicity tests, RAMS amplifications of genomic DNA, species-specific PCR and PCR-RFLP of the IGS region from rDNA were used.
Materials and methods
Isolates and cultural conditions
Forty-two isolates were obtained in 2011 from 37 root and 160 stem samples of 160 trees showing vascular discoloration from various olive cultivars in eight regions in Tunisia, including Monastir (Sahline, Boumerdes, Zarmedine), Kasserine, Sousse (Chott-Mariem), Sfax (Sidi-Bouakkazine), Mahdia, Sidi-Bouzid (Souk-jdid), Zaghouan (Nadhour) and Kairouan (Chbika) (). The samples were collected from different orchards and carried separately to the laboratory for isolation. Infected plant tissues were rinsed twice in sterilized distilled water, and then disinfected with 95% ethanol for 2 min prior to being cut aseptically into 0.5 to 2-cm pieces. Four to five pieces from each sample were placed onto each potato dextrose agar (PDA) plate. After 2 weeks of incubation at 25 °C, isolates were identified based on colony morphology and microscopic characteristics of conidia and microsclerotia production (Triki et al. Citation2006). Fungal isolates derived from single-spore cultures were obtained using the serial spore dilution method (Choi et al. Citation1999). Briefly, 1 mL of each serial dilution was mixed with melted nutrient agar and transferred to sterile plates. The plates were incubated at 25 °C in the dark and then visualized under the microscope to locate the germinating spores. These latter were cut from the medium and transferred to new PDA plates for development of colonies. The pure cultures were maintained on PDA slants at 4 °C (Choi et al. Citation1999). The isolates were cultured in potato dextrose broth (PDB) on a rotary shaker (150 rpm) at 25 °C. The fungal mycelium was recovered by filtering and lyophilized and stored at −20 °C until used.
Table 1. Verticillium dahliae isolates collected from olive tree (Olea europea) in Tunisia.
Pathogenicity tests
The pathogenicity of the isolates was tested using a root dipping method on 2-year-old olive trees ‘Chemlali’ according to Rodriguez Jurado et al. (Citation1993). Mycelium was recovered by scraping aseptically the surface of 10-day-old cultures on PDA and transferring into flasks containing sterile PDB medium. The liquid cultures were then incubated for 7 days at 22 °C in the dark to obtain spores. The conidial suspension was adjusted to 106 conidia/mL. Plant roots were washed, dried and dipped for 1 h in the conidial suspension. After inoculation, olive trees were transplanted into polyethylene pots containing a sterile substrate (peat:sand, 1:1 v/v). Plants were placed in a growth chamber set at constant 23 ± 2 °C with a 12 h photoperiod provided by fluorescent lights of 216–270 µEm−2 s−1. The pots were arranged in a completely randomized block design with three replicates per isolate and three control plants dipped in sterile water. The experiment was repeated twice. Disease severity was assessed visually each week, starting at 15 days after inoculation. A scale from 0 to 4 was used according to the percentage of affected plant tissue (0 = healthy plant; 1 = ≤ 33% affected tissue; 2 = 34–66% affected tissue; 3 = 67–99%; 4 = dead plant). Estimation of the area under disease progress curve (AUDPC) was calculated as described previously by Campbell & Madden (Citation1990). Statistical analysis of variance was performed using GraphPad Prism (GraphPad Software) to determine the variability among the 42 isolates tested.
Genomic DNA extraction
Forty-two V. dahliae isolates were grown on PDA at 25 °C for 1 week in the dark. After incubation, mycelia were scraped off the surface of colonies using a sterile scalpel and ground in liquid nitrogen with sterilized sea sand. DNA was then extracted from 100 mg of ground mycelium using the phenol/chloroform method. After centrifugation at 10 000 rpm for 10 min, the pelleted DNA was washed with 70% ethanol, air dried and resuspended in TE buffer. DNA elution was quantified using a Nanodrop, ND-1000 spectrophotometer (Thermo Fisher Scientific). Aliquots of samples were also analysed on a 0.7% agarose gel to check DNA quality (Raeder and Broda Citation1985).
PCR assays
All isolates were characterized using ITS-based specific primers for V. dahliae that yield a 334 bp amplicon (Robb et al. Citation1994). PCR reactions were performed in 25 µL final volume containing 250 µM dNTPs mix, 1 μM of each primer, 2.5 mM MgCl2, 1 U Taq DNA polymerase and 50 ng of fungal DNA. Amplification of the IGS region was carried out using primers CNL12 and CNS1 (). PCR reactions of the IGS region were carried out in 25 μL reaction mixture containing 1 × PCR buffer (including 1.5 mM MgCl2), 250 μM dNTPs mix, 0.5 μM of each primer, 1 U Taq DNA polymerase and 10 ng of the template DNA. Optimal amplifications for the IGS region were obtained with an initial denaturation at 95 °C for 2 min followed by 35 amplification cycles (denaturation, 95 °C for 35 s, annealing, 58 °C for 55 s, and extension, 72 °C for 45 s), and a final extension at 72 °C for 10 min. PCR products were digested without prior purification using restriction endonucleases EcoRI and RsaI (England Biolabs). Amplicons and their restriction fragments were visualized on 1.5% agarose gels containing 0.5 µg mL−1 ethidium bromide at 80 volts for 30 min along a 1-kb DNA ladder (Gibco BRL).
Table 2. List of amplified genomic regions and primers.
RAMS analysis
Five primers, previously described by Hantula et al. (Citation1996), were selected for RAMS amplifications (). Amplifications were performed in a total reaction volume of 25 μL containing 1 × PCR buffer (including 1.5 mM MgCl2), 100 μM each dNTP, 2 μM primer, 1 U Taq DNA polymerase and 10 ng of DNA. The thermal cycler was programmed with temperature profiles as described in Hantula et al. (Citation1998). PCR amplifications were carried out in duplicate for each isolate.
Data analysis
Comparison of each banding pattern generated by all the different primers was noted on the basis of the presence [1] or absence [0] of polymorphic markers. Scoring of RAMS amplification was performed manually from gel pictures. After comparison of the results of repeated experiments, only 21 polymorphic and reproducible bands were included in the analysis. The relationships between all the isolates could be represented by a similarity matrix. Scoring of RAMS amplification was performed visually from photographic prints. Data were analysed using NTSYSpc version 2.0 to generate a genetic similarity matrix using the genetic distance coefficient, and a dendrogram was generated using the neighbour-joining method (Rohlf Citation1988).
Results
Morphological and microscopic identification
Microscopic examination of the isolates revealed the presence of hyaline conidiophores with verticillate branches, 2–4 phialides at each node. For each isolate, the measurement of 10 conidia per isolate indicated a size of 2.5–5 µm × 1.25–2.5 µm (A). Melanized microsclerotia were seen 7–10 days after incubation (B). Colonies were generally white or white to grey () and isolates were divided into two morphotypes based on colony colour and texture: milky white and flocculose to slightly flocculose mycelium (MW) (C) (36 isolates) and greyish-white surface and slightly flocculose texture (GW) (D) (six isolates) (). All of the isolates used in this study yielded a specific 334 bp band using the primer pair Vd1/Vd2 ().
Fig. 1 (Colour online) Morphological characteristics of different V. dahliae morphotypes cultured on PDA for 15 days at 25 °C. A: Morphotype 1 with milky white and flocculose mycelium; B: Morphotype 2 with greyish-white surface; C: Morphology of conidiophores, phialides and conidia of V. dahliae; D: pure culture of V. dahliae.
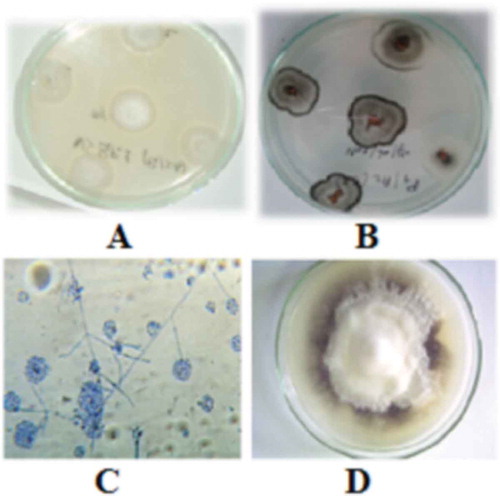
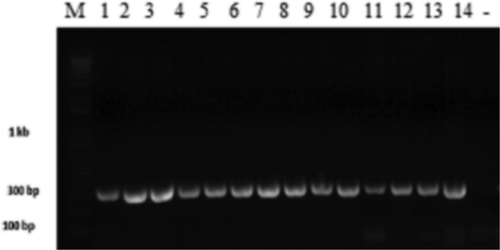
Fig. 2 Gel electrophoresis of the amplified rDNA internal transcribed spacer (ITS) specific region of Verticillium dahliae isolates. Lanes 1–16 are loaded as follows: DNA size marker, PCR products of Vsb 1.2, VSBP7, Vms 3.3, Vmh3.2, VK1.11, Vk1.3, Vcm2, V.hmd, VSB1.1, V30, Vd4, Vz17, VS 1.1G, Vms 3.2, and negative control.
Pathogenicity tests
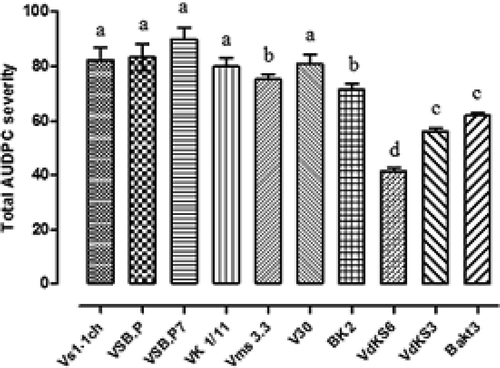
All of the V. dahliae isolates used in this study were pathogenic on olive. Wilt symptoms appeared 1 month after inoculation. Leaves became chlorotic and gradually turned brown with a progressive rolling inwards. Two months after inoculation, the disease severity reached 40% for almost all of the isolates. A statistical analysis was performed and the differences among isolates were statistically significant (P < 0.05). AUDPC values ranged between 41.25% and 89.12% (). Isolates such as VSBP7 from Sidi bouzid, VMS 3.3 from Monastir and Vk1/11 from Kasserine had the highest level of aggressiveness (P < 0.05), whereas isolates Vdks3, Vdks6 from Kasserine and Bakt3 from Sfax, were less aggressive.
PCR-RFLP analysis of the IGS region
A single PCR product, ranging from 1.7 to 2.1 kb in size, was obtained from the 42 isolates. The variation in size of the PCR product may be due to the frequent insertion of introns of variable sizes within the small unit and the large unit gene regions. Digestion of IGS sequences with EcoRI and RsaI generated 2–3 restriction fragments for each enzyme (). The digestion profile obtained by EcoRI varied among the tested isolates. Interestingly, EcoRI had no restriction site in one isolate (Vmh3.2). Digestion of the IGS sequences with RsaI generated similar patterns (1200 and 800 bp) for 40 isolates, but two isolates, Vs1.1ch and Vs1.2ch, were different and shared the same banding pattern (1100, 850 and 150 bp).
Table 3. Banding patterns of the amplified rDNA intergenic spacer (IGS) region after digestion with RsaI and EcoRI.
RAMS analysis
RAMS analysis using three repeat primers showed variable banding patterns among the isolates. Nine (2200, 1400, 1200, 1100, 1000, 650, 550, 500 and 350 bp), eight (2000, 1200, 950, 850, 800, 550, 500 and 400 bp) and four bands (2500, 2400, 700 and 500) were produced using primers CGA, CCA and GT, respectively (, ). All three sets of RAMS primers provided reproducible data in two separate experiments. When data from the RAMS experiments were combined, a total of 21 markers were identified. Each marker was named according to its size (). The banding pattern in the V. dahliae isolates was categorized as moderate due to the relatively low number of amplification products (). The number of fragments amplified was always greater when CGA, rather than CCA or GT, was used as a primer and this occurred with all of the tested isolates. The observed variation was analysed according to the presence or absence of specific bands. Using RAMS primer GT, only a few polymorphic markers were observed among the tested isolates, particularly DNA fragments 2500, 2400 and 700 bp, which were only present in isolates Vd2, Vk6/11, Vk3/11, Vs3.1F and Vs3.3F (). Using CCA primer, only one band of 850 bp (zarmdine), 900 bp (Vdks5) or 800 bp (Vs3.1F and Vs3.3F) was observed. ACA and GAAA primers did not generate reproducible banding profiles from the isolates.
Fig. 4 Random amplified microsatellite (RAMS) pattern of Verticillium dahliae isolates using CGA primer. Lanes 1–15 are loaded with PCR products of VDKS5, Vd4, BAKT3, VS3.3F, VS1.2 T, VMS3.2, VMS2, VDKS6, VDKS3, VK1.2, Vd2, VK6.11, VFA2, BAKT11 and BAKT7, respectively. DNA size marker is indicated by M.
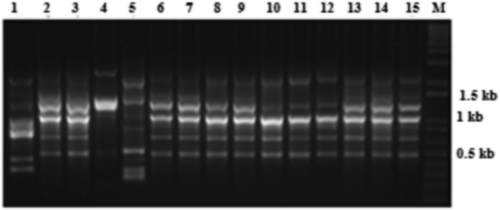
Fig. 5 Random amplified microsatellite (RAMS) pattern of Verticillium isolates using CCA primer. Lanes 1–21 are loaded with PCR products of Vsh1.2, VSBP7, BK2, Vmh3.2, VK1.11, Vk1.3, Vcm2, VS1.1ch, VSB1.1, Vsh, V30, Vd3, Vd4, Vz17, VSB1.4, VHmd, Vd2, VDOR, VS1.2ch, VS1.1 G and BAKT7, respectively. DNA size marker is indicated by M.
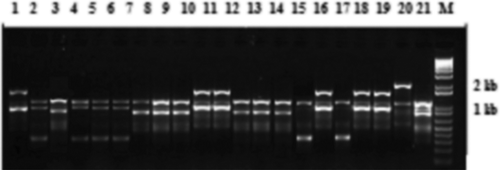
Table 4. RAMS banding patterns of the tested V. dahliae isolates using primers GT, CCA and CGA.
Phylogenetic analysis
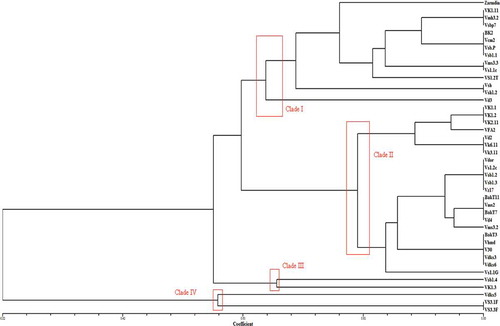
The UPGMA analysis identified four major clades based on similarity coefficient values of approximately 0.22–1 (). The genetic similarity within these clades was 65% for clade I, 80% for clade II, 66.9% for clade III and 57% for clade IV. Each major clade contained a combination of isolates from different regions, indicating that genotypes did not group according to their geographic origin. Clades I and II, the most diverse geographically, had 12 isolates from Sfax, six from Sidi Bouzid, six from Kasserine, six from Monastir and three from Zaghouane. Clade IV had only two isolates from Sfax. There was no clear evidence for geographic clustering of the tested isolates.
Discussion
The use of RAMS and PCR-RFLP of the IGS region revealed that V. dahliae isolates from diseased olive trees in Tunisia were grouped according to their virulence phenotype, regardless of their morphological features or geographic origin. Morphological characterization of V. dahliae recovered from infected olive trees indicated the presence of two main morphotypes, with the major one having a milky white mycelium, that gradually darkens with microsclerotia formation. Pegg & Brady (Citation2002) reported a similar morphological diversity when studying mycelial growth of V. dahliae on artificial media, and indicated that temperatures over 28 °C can lead to a higher frequency of morphological mutants (Pegg & Brady Citation2002). Agrochemicals have also been shown to act as mutagens on Verticillium sp. (Galperina Citation1990). The distribution of these two morphotypes in the dendrogram () showed no obvious correlation, with isolates randomly grouped regardless of their morphological features. For instance, four isolates (Vms 3.3, Vsb 1.1, Vsb.P and VSB.P7) out of six, which belong to the morphotype WG, were assigned to clade I with 10 isolates from the morphotype MW. This result suggests that there was admixture between the two observed morphotypes.
The IGS region appeared to be moderatly divergent both in length and restriction patterns among the tested V. dahliae isolates. Coupled with pathogenicity data, the IGS-RFLP failed to group the isolates according to their level of aggressiveness. This result supported previous findings that IGS region is not a reliable marker for intraspecific classification of V. dahliae isolates according to their pathogenicity patterns. Different overall IGS sizes were also reported by Pramateftaki et al. (Citation2000), with an IGS size of 1.6 kb in 31 V. dahliae strains, and 1.7 kb in another set of 10 V. dahliae isolates. In the same study, examination of 12 V. dahliae isolates with different morphology, pathogenicity, geographic and host origins and another 29 V. dahliae isolates with similar morphology and origin showed no association of rDNA size with any of these characteristics. Results from the present study indicate that correlations between intraspecific V. dahliae groups and their geographic origins seem unlikely, given that isolates from different localities showed similar restriction patterns when EcoRI or RsaI were used. In a study conducted by Otero et al. (Citation2004), although the IGS size was constant, variability among the tested isolates from the same host species was detected when RsaI and DraI were used, without any obvious correlations with virulence or geographic origin. The present study revealed moderate variability in isolates from olive, with different haplotypes.
RAMS provided a differentiation of the tested isolates, and all isolates had polymorphic amplification products when the three RAMS repeat primers (CCA, CGA and GT) were used (, ). In fact, RAMS analysis using these primers resulted in banding pattern variations between the isolates from diseased olive trees, indicating a relatively high degree of genetic variation among the isolates and that these microsatellite motifs exist abundantly in the genome of the isolates used in this study. Pathogenicity tests revealed a correlation between the genetic variation observed by RAMS and pathogenicity patterns of the isolates. For example, VSB.P, VSB.P7, BK2, Vms 3.3, Vs 1.1ch and Vk1/11, which are the most pathogenic isolates on olive, were assigned to clade I, while isolates with moderate pathogenicity were mostly assigned to clades II, III and IV. In contrast, V30, which is a highly aggressive isolate, formed a cluster with the weakly aggressive isolates Vdks3, Vdks6 and BAKT3.
Results from the present study suggest that diversity within this population is not correlated with geographic origin. For instance, in the phylogenetic analysis, three isolates from Sidi Bouzid (VSB.P, VSBP 7, VSB 1.1), three from Monastir (Vsh, Vsh 1.2, Vms 3.3) and three from Sfax (Vs 1.2T, Vs 1.1ch, BK2) formed a cluster, although they came from various locations. Furthermore, isolates Vsb 1.4 and VK 1.3 are from different locations, but formed an individual cluster (). All these observations indicated a lack of clustering of isolates according to their virulence patterns. The number of RAMS markers observed among V. dahliae isolates in the present study was higher than other studies in Gremmeniella abietina (Hantula & Müller Citation1997), Peridermium pini (Hantula et al. Citation1998) and Sphaeropsis sapinea (Burgess et al. Citation2001), suggesting that either the genome size is larger or the frequency of microsatellites with particular CGA-motifs is higher in Verticillium than in other fungi.
The relatively high number of RAMS markers with primer CGA in this study may be indicative of genetic variation among V. dahliae isolates from different olive growing regions, which would suggest the possibility that V. dahliae was introduced into different regions of Tunisia from different source populations rather than having originated from a common Tunisian ancestor. Compared with other V. dahliae studies with isolates collected from different hosts and locations, the level of genetic variation reported in this study would be similar to that reported by Collado-Romero et al. (Citation2006), who conducted an analysis of V. dahliae isolates from artichoke, olive, cotton and soil using AFLP, with a maximum genetic variation of 88% for all isolates. Erdogan et al. (Citation2013) also reported a similar result with AFLP analysis of V. dahliae isolates collected from very close geographic regions in Western Turkey where cotton is extensively grown, with a maximum genetic variation of 79%.
Using RAPD, phylogenetic analysis using 27 V. dahliae isolates from olive in neighbouring Kabylia and the north-western part of Algeria showed a low level of genetic variation, with isolates tending to cluster together in the same group (Bellahcene et al. Citation2005). In Morocco, an earlier RAPD study of a collection of V. dahliae isolates infecting olive trees, identified groups based on their local geographic origin (Cherrab et al. Citation2000). In addition to natural genetic variation, both human and environmental factors may play a role in differential selection of V. dahliae strains in different regions of Tunisia. The spreading of vegetative material over a wide geographic area can also be another factor in shaping the population structure of V. dahliae (Thanassoulopoulos Citation1993). In fact, olive is extensively grown in the tested regions, and growers use olive orchards for production of solanaceous species that are susceptible to V. dahliae. These management practices may lead to selection pressure in the pathogen population, thereby contributing to the development of new V. dahliae pathotypes (Lopez-Escudero et al. Citation2010). Cross-pathogenicity of V. dahliae among different crops has been shown (Alkher et al. Citation2009), and intensive olive cropping along with the introduction of imported olive cultivars may add to the emergence of new pathotypes. Such variation, in parallel with recent epidemics, may result in resistance breakdown in the main olive cultivars, namely ‘Chemlali’ and ‘Chetoui’, which have been cultivated for approximately one century. Thus, continuous olive breeding aimed at developing cultivars resistant to newly emerging pathotypes is needed, and screening for disease resistance should include isolates that would reflect genetic diversity found in this study. Hence, understanding the genetic diversity of V. dahliae on olive in Tunisia will aid in future disease management strategies of Verticillium wilt in this and neighbouring countries. Future studies to improve the understanding of genetic variation in the local populations of V. dahliae should be conducted using additional, co-dominant molecular markers and more isolates from different hosts. The use of RAMS and IGS-RFLP provided the first data on the structure of V. dahliae populations now present in olive trees in Tunisia, and showed a moderate level of their genetic variation which is correlated with the virulence phenotype, but with no obvious clustering of isolates according to their geographic origin or morphological features.
Acknowledgements
This study was supported in part by grants from the PESTOLIVE project in Tunisia to M.A. Triki and Research Laboratory APREGO, the research unit of toxicology – environmental microbiology and health to R. Gdoura, and NSERC to F. Daayf. We thank Mr Lorne R. Adam, Department of Plant Science, University of Manitoba, Winnipeg, for his technical help.
References
- Alkher H, El Hadrami A, Rashid KY, Adam LR, Daayf F. 2009. Pathogenic variation of Verticillium dahliae after serial passages through potato and sunflower. Can J Plant Pathol. 31:427–438. doi:10.1080/07060660909507617
- Appel D, Gordon TR. 1995. Intraspecific variation within populations of Fusarium oxysporum based on RFLP analysis of the intergenic spacer region of the rDNA. Exp Mycol. 19:120–128. doi:10.1006/emyc.1995.1014
- Bainbridge BW. 1994. Modern approaches to the taxonomy of Aspergillus. In: Powell KA, Peberdy J and Renwick A, editors. Genetics and physiology of Aspergillus. New York: Plenum Press; 291–301.
- Barbara DJ, Clewes E. 2003. Plant pathogenic Verticillium species: how many of them are there?. Mol Plant Pathol. 4:297–305. doi:10.1046/j.1364-3703.2003.00172.x
- Bellahcene M, Assigbetsi K, Fortas Z, Geiger JP, Nicole M, Fernandez D. 2005. Genetic diversity of Verticillium dahliae isolates from olive trees in Algeria. Phytopathol Mediter. 74:266–274.
- Bhat RG, Subbarao KV. 1999. Host range specificity in Verticillium dahliae. Phytopathology. 89:1218–1225. doi:10.1094/PHYTO.1999.89.12.1218
- Bilodeau GJ, Koike ST, Uribe P, Martin FN. 2012. Development of an assay for rapid detection and quantification of Verticillium dahliae in soil. Phytopathology. 102:331–343. doi:10.1094/PHYTO-05-11-0130
- Børja I, Solheim H, Hietala AM, Fossdal CG. 2006. Etiology and real time polymerase chain reaction based detection of Gremmeniella and Phomopsis associated disease in Norway spruce seedling. Phytopathology. 96:1305–1314. doi:10.1094/PHYTO-96-1305
- Burgess T, Wingfield MJ, Wingfield BW. 2001. Simple sequence repeat markers distinguish among morphotypes of Sphaeropsis sapinea. Appl Environ Microbiol. 67:354–362. doi:10.1128/AEM.67.1.354-362.2001
- Campbell CL, Madden LV. 1990. Introduction to plant disease epidemiology. New York: John Wiley and Sons; p. 532.
- Cherrab M, Serrhini MN, Charest PM. 2000. Characterization of Moroccan isolates of Verticillium dahliae Kleb. Using RAPD markers. J Phytopathol. 148:243–249. doi:10.1046/j.1439-0434.2000.00500.x
- Choi YW, Hyde KD, Ho W. 1999. Single spore isolation of fungi. Fungal Diversity. 3:29–38.
- Cohen S, Allasia V, Venard P, Notter S, Vernière CH, Panabières F. 2003. Intraspecific variation in Phytophthora citrophthora from citrus trees in eastern Corsica. Eur J Plant Pathol. 109:791–805. doi:10.1023/A:1026190318631
- Collado-Romero M, Mercado-Blanco J, Olivares-García C, Valverde-Corredor A, Jiménez-Díaz RM. 2006. Molecular variability within and among Verticillium dahliae vegetative compatibility groups determined by fluorescent amplified fragment length polymorphism and polymerase chain reaction markers. Phytopathology. 96:485–495. doi:10.1094/PHYTO-96-0485
- Daayf F, Nicole M, Geiger JP. 1995. Differentiation of Verticillium dahliae populations on the basis of vegetative compatibility and pathogenicity on cotton. Eur J Plant Pathol. 101:69–79. doi:10.1007/BF01876095
- Dobinson KF, Harrington MA, Omer M, Rowe RC. 2000. Molecular characterization of vegetative compatibility group 4A and 4B isolates of Verticillium dahliae associated with potato early dying. Plant Dis. 84:1241–1245. doi:10.1094/PDIS.2000.84.11.1241
- El-Bebany AF, Alkher H, Adam LR, Daayf F. 2013. Vegetative compatibility of Verticillium dahliae isolates from potato and sunflower using nitrate-nonutilizing (nit) mutants and PCR-based approaches. Can J Plant Pathol. 35:1–9. doi:10.1080/07060661.2012.702128
- El-Bebany AF, Henriquez MA, Badawi M, Adam L.R., El Hadrami A, Daayf F. 2011. Induction of putative pathogenicity-related genes in Verticillium dahliae in response to elicitation with potato root extracts. Environ Exp Bot. 72:251–257. doi:10.1016/j.envexpbot.2011.03.012
- El-Bebany AF, Rampitsch C, Daayf F. 2010. Proteomic analysis of the phytopathogenic soilborne fungus Verticillium dahliae reveals differential protein expression in isolates that differ in aggressiveness. Proteomics. 10:289–303. doi:10.1002/pmic.200900426
- El Hadrami A, Adam LR, Daayf F. 2011. Biocontrol treatments confer protection against Verticillium dahliae infection of potato by inducing anti-microbial metabolites. Mol. Plant-Microbe Interact. 24:328–335. doi:10.1094/MPMI-04-10-0098
- Erdogan O, Nemli S, Oncu T, Tanyolac B. 2013. Genetic variation among pathotypes of Verticillium dahliae Kleb. from cotton in western Turkey revealed by AFLP. Can J Plant Pathol. 35:354–362. doi:10.1080/07060661.2013.809790
- Food and Agriculture Organization of the United Nations. 2008. FAOSTAT online database. Available from: http://www.fao.org/corp/statistics/en/
- Galperina MI. 1990. Genetic effects of some pesticides on Verticillium spp. Proceedings of the Fifth International Verticillium symposium, June 25–30, Leningrad, USSR; p. 101.
- Hantula J, Dusabenyagasani M, Hamelin RC. 1996. Random amplified microsatellites (RAMS) ? a novel method for characterizing genetic variation within fungi. Forest Pathology. 26:159–166. doi:10.1111/j.1439-0329.1996.tb00720.x
- Hantula J, Müller M. 1997. Variation within Gremmeniella abietina in Finland and other countries as determined by Random Amplified Microsatellites (RAMS). Mycol Res. 101:169–175. doi:10.1017/S0953756296002225
- Hantula J, Niemi EM, Kaitera J, Jalkanen R, Kurkela T. 1998. Genetic variation of the resin top fungus in Finland as determined by random amplified microsatellites (RAMS). Forest Pathology. 28:361–372. doi:10.1111/j.1439-0329.1998.tb01190.x
- Jabnoun-Khiareddine H, Daami-Remadi M, Hibar K, Ayed F, El Mahjoub M. 2006. Pathogenecity of Tunisian Isolates of Three Verticillium Species on Tomato and Eggplant. Plant Pathol J. 5:199–207. doi:10.3923/ppj.2006.199.207
- Jurado D, López MA, Rapoport HF, Diaz RM. 1993. Present status of verticillium wilt of olive in Andalucía (southern Spain). EPPO Bull. 23:513–516. doi:10.1111/j.1365-2338.1993.tb01362.x
- Klosterman SJ, Atallah ZK, Vallad GE, Subbarao KV. 2009. Diversity, pathogenicity and management of Verticillium species. Annu Rev Phytopathol. 47:39–62. doi:10.1146/annurev-phyto-080508-081748
- Korolev N, Pérez-Artés E, Mercado-Blanco J, Bejarano-Alcázar J, Rodríguez-Jurado D, Jiménez-Díaz RM, Katan T, Katan J. 2008. Vegetative compatibility of cotton-defoliating Verticillium dahliae in Israel and its pathogenicity to various crop plants. Eur J Plant Pathol. 122:603–617. doi:10.1007/s10658-008-9330-1
- Lopez-Escudero FJ, Mercado-Blanco J, Roca JM, Valverde-Corredor A, Blanco-López MA. 2010. Verticillium wilt of olive in the Guadalquivir Valley (southern Spain): relations with some agronomical factors and spread of Verticillium dahliae. Phytopathol Med. 49:370–380.
- Mahuku GS, Henríquez MA, Munõz J, Buruchara R. 2002. Molecular Markers Dispute the Existence of the Afro-Andean Group of the Bean Angular Leaf Spot Pathogen,Phaeoisariopsis griseola. Phytopathology. 92:580–589. doi:10.1094/PHYTO.2002.92.6.580
- Mishra P, Fox ROLAND T V., Culham A. 2003. Inter-simple sequence repeat and aggressiveness analyses revealed high genetic diversity, recombination and long-range dispersal in Fusarium culmorum. Ann Appl Biol. 143:291–301. doi:10.1111/j.1744-7348.2003.tb00297.x
- Morton A, Carder JH, Barbara DJ. 1995. Sequences of the internal transcribed spacers of the ribosomal RNA genes and relationships between isolates of Verticillium albo-atrum and V. dahliae. Plant Pathol. 44:183–190. doi:10.1111/j.1365-3059.1995.tb02727.x
- Nazar RN, Hu X, Schmidt J, Culham D, Robb J. 1991. Potential use of PCR-amplified ribosomal intergenic sequences in the detection and differentiation of Verticillium wilt pathogens. Physiol Mol Plant Pathol. 39:1–11. doi:10.1016/0885-5765(91)90027-F
- Otero L, Ducasse D, Miller RNG. 2004. Variability in ribosomal DNA genic and spacer regions in Verticillium dahliae isolates from different hosts. Fitopatol Brasil. 29:441–446. doi:10.1590/S0100-41582004000400015
- Papaioannou IA, Dimopoulou CD, Typas MA. 2013. Structural and phylogenetic analysis of the rDNA intergenic spacer region of Verticillium dahliae. FEMS Microbiol Lett. 347:23–32. doi:10.1111/1574-6968.12215
- Pegg GF, Brady BL. 2002. Verticillium wilts. Wallingford: CAB International.
- Pramateftaki PV, Antoniou PP, Typas MA. 2000. The complete DNA sequence of the nuclear ribosomal RNA gene complex of Verticillium dahliae: intraspecific heterogeneity within the intergenic spacer region. Fungal Genet Biol. 29:19–27. doi:10.1006/fgbi.1999.1178
- Qin QM, Vallad GE, Wu B.M., Subbarao KV. 2006. Phylogenetic analyses of phytpathogenic isolates of Verticillium spp. Phytopathology. 96:582–592. doi:10.1094/PHYTO-96-0582
- Raeder U, Broda P. 1985. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1:17–20. doi:10.1111/j.1472-765X.1985.tb01479.x
- Ratanacherdchai K, Waang HK, Lin FC, Soytong K. 2010. ISSR for comparison of cross-inoculation potential of Colletotrichum capsici causing chilli anthracnose. Afr J Microbiol Res. 4:76–83.
- Robb J. 2007. Verticillium tolerance: resistance, susceptibility or mutualism?. Can J Bot. 85:903–910. doi:10.1139/B07-093
- Robb J, Hu X, Platt H, Nazar R. 1994. PCR assay for the detection and quantification of Verticillium species in potato. In: Schots A, Dewey FM, Oliver R, editors. Modern detection assay for plant pathogenic fungi: identification, detection and quantification. Oxford: CAB International; p. 83–90.
- Rohlf JF. 1988. NTSYS-pc: numerical taxonomy and multivariate analysis system. Version 1.80. Exeter Software. Setauket, NY
- Thanassoulopoulos CC. 1993. Spread of verticillium wilt by nursery plants in olive groves in the Halkidiki area (Greece). EPPO Bull. 23:517–520. doi:10.1111/j.1365-2338.1993.tb01363.x
- Tooley PW, O’Neill NR, Goley ED, Carras MM. 2000. Assessment of diversity in Claviceps africana and other claviceps species by RAM and AFLP analyses. Phytopathology. 90:1126–1130. doi:10.1094/PHYTO.2000.90.10.1126
- Triki MA, Hassaïri A, Mahjoub M. 2006. Premières observations de Verticillium dahliae sur olivier en Tunisie. EPPO Bull. 36:69–71. doi:10.1111/j.1365-2338.2006.00941.x
- Zhou S, Smith DR, Stanosz GR. 2001. Differentiation of Botryosphaeria species and related anamorphic fungi using inter simple or short sequence repeat (ISSR) fingerprinting. Mycol Res. 105:919–926. doi:10.1016/S0953-7562(08)61947-4
- Zietkiewicz E, Rafalski A, Labuda D. 1994. Genome fingerprinting by simple sequence repeat (SSR) - anchored polymerase chain reaction amplification. Genomics. 20:176–183. doi:10.1006/geno.1994.1151