Abstract
Throughout the plant disease cycle, biotrophic fungal pathogens must obtain host-derived carbon molecules to act as building blocks and sources of energy. Gaining access to these resources requires biotrophic fungi to breach plant cell walls without eliciting substantial plant defences. The plant cell wall is composed mostly of glucose- and xylose-based polysaccharides, which can support fungal growth. Thus, fungi can acquire carbon compounds through the targeted depolymerization of specific wall components. When the plant cell wall is breached, biotrophs redirect photoassimilates, increase sink strengths, and express invertases and transporters to acquire carbon compounds. Transitions in enzyme and transporter expression during pathogenesis must be tightly controlled to ensure a continued supply of carbon compounds. This review describes carbon acquisition and metabolism, including regulation of available carbon source utilization mechanisms such as carbon catabolite repression. While carbon acquisition has been extensively studied in the ascomycetes, the mechanisms used by biotrophic fungi to acquire carbon during pathogenesis are poorly understood. Furthermore, the relationship between plant cell wall-degrading enzymes and carbon acquisition in biotrophic fungal pathogens is not well characterized. As such, this review summarizes the current knowledge of carbon source utilization by fungal pathogens, with an emphasis on research involving the corn smut pathogen Ustilago maydis, and provides a basis from which to extend our knowledge in this key area of fungal plant pathogenesis.
Résumé
Durant le cycle de la maladie de la plante, les agents pathogènes fongiques biotrophes doivent obtenir de leur hôte les molécules de carbone qui lui serviront d’éléments constitutifs et de source d’énergie. Pour accéder à ces ressources, le champignon biotrophe doit percer la membrane cellulaire sans pour cela déclencher une réaction de défense vigoureuse de la plante. La membrane cellulaire est composée principalement de polysaccharides constitués de glucose et de xylose qui peuvent supporter la croissance des champignons. Ainsi, les champignons peuvent obtenir les composés de carbone par la dépolymérisation ciblée de composants précis de la membrane. Lorsque la membrane de la plante est percée, l’agent biotrophe redirige les photoassimilats, accroît l’intensité du puits et extrait les invertases et les transporteurs afin d’obtenir les composés de carbone requis. Les transitions relatives à l’expression des enzymes et des transporteurs durant la pathogenèse doivent être étroitement contrôlées afin d’assurer un approvisionnement continu en carbone. Cet article décrit l’acquisition et le métabolisme du carbone, y compris la régulation des mécanismes d’utilisation des sources de carbone disponibles, telle la répression catabolique du carbone. Tandis que l’acquisition du carbone a été étudiée extensivement chez les ascomycètes, les mécanismes utilisés par les champignons biotrophes pour acquérir du carbone durant la pathogenèse sont mal compris. De plus, la relation qui existe entre les enzymes qui dégradent la membrane cellulaire et l’acquisition du carbone par les agents pathogènes fongiques biotrophes n’est pas bien caractérisée. À ce titre, l’article résume les connaissances actuelles sur l’utilisation des sources de carbone par les agents pathogènes fongiques, et ce, en mettant l’accent sur la recherche concernant Ustilago maydis, l’agent pathogène du charbon du maïs, et pose une base à partir de laquelle nous pourrons étendre nos connaissances dans ce domaine clé de la pathogenèse fongique chez les plantes.
Introduction
Carbon acquisition is a fundamental requirement for fungal cell development and proliferation as carbon source molecules taken up by fungi become the main structural component of cell walls, proteins and nucleic acids. Carbon compounds are also oxidized to produce energy and are involved in cellular signalling pathways in fungi. If the fungus is a saprobe or necrotroph, these essential carbon compounds come from the degradation of organic matter or dead plant material, respectively; however, biotrophs must obtain carbon compounds from their hosts without killing them (Divon & Fluhr Citation2007). This means that biotrophic fungi must breach cell walls and access sufficient carbon sources in the plant to enable fungal growth, but do so in a manner that is not so damaging as to elicit dramatic protection responses from the plant, such as a hypersensitive response (Heath Citation1998). Surprisingly, little is known about how fungal biotrophs modulate the process of carbon acquisition. In contrast, there is a wealth of knowledge regarding carbon acquisition and carbon catabolite repression (CCR) in several model ascomycete fungi and substantial information on the biotechnological use of cell wall-degrading enzymes from fungi (reviewed in Ruijter & Visser Citation1997; de Vries Citation2003; van den Brink & de Vries Citation2011; Brown et al. Citation2014). This review seeks to examine how carbon acquisition is modified by biotrophic fungi during pathogenic development in plants. The model biotroph Ustilago maydis is used to frame the discussions relating to carbon sources in plant tissues, enzymatic digestion of plant barriers, carbon uptake and metabolism, carbon catabolite repression, and how all of this relates to biotrophic plant pathogenesis and symptom development of corn smut.
Plant cell wall degradation and carbon acquisition by fungi
Fungi must obtain carbon from their local environment and, in the case of biotrophic pathogens, this environment is a living host plant. Carbon compounds, which are abundant in plant cell walls, can be accessed by fungi through the secretion of cell wall-degrading enzymes (CWDEs). During entry into and growth within the plant, CWDEs are released by the fungus, facilitating cell wall breakdown and hyphal extension between and, in the case of U. maydis, within plant cells (Banuett & Herskowitz Citation1996; reviewed in Rytioja et al. Citation2014). However, the free simple sugars and more complex polymers available inside plant cells and in the plant apoplast may be more readily metabolized by fungi. Therefore, the mechanisms by which fungi acquire carbon molecules may change as infection proceeds. Carbon sources that are circulating through the plant or present in cells can be processed by invertases and taken up by fungal transporters (reviewed in Doidy et al. Citation2012; Tauzin & Giardina Citation2014). To provide context for discussing mechanisms of carbon acquisition by biotrophic fungi, and the enzymes involved, one must consider the nature of the carbon sources in the plant.
Plant cell walls provide support and rigidity as well as protection from invading microorganisms. The structural complexes that form plant cell walls include cellulose, hemicellulose, pectin and lignin (reviewed in Mäkelä et al. Citation2014; Rytioja et al. Citation2014). As an example, the secondary wall of a monocotyledonous plant such as corn is often generalized as consisting of 35–45% cellulose, 40–50% hemicellulose, 0.1% pectin and 20% lignin, though the exact composition of the plant cell wall can vary with growth stage (Vogel Citation2008; Benatti et al. Citation2012). Celluloses are comprised of 1,4-β-linked glucose units that are organized into linear microfibrils, which are crystalline structures held together by intermolecular hydrogen bonds that wrap around cells (Kolpak & Blackwell Citation1976). Hemicelluloses, another abundant plant polysaccharide, include xylan, mannan or xyloglucan, which contain linked monosaccharide pentose or hexose molecules such as xylose, mannose and glucose, respectively (reviewed in Mäkelä et al. Citation2014). Other monomers such as galactose, arabinose or glucuronic acid can also be attached to the main hemicellulose backbone (van den Brink & de Vries Citation2011). The less abundant pectin is a heterogenic polymer mostly comprised of α-1,4-linked galacturonic acid residues which provides additional cross-link support between celluloses and hemicelluloses (reviewed in van den Brink & de Vries Citation2011). Finally, lignins are complex polymers that provide added support to cell walls through cross-linking of celluloses, hemicelluloses and pectins (Grabber et al. Citation2000). A number of other reviews have provided excellent diagrams, outlining the structure of plant cell walls and their degradation by enzymes. Readers are referred to Aro et al. (Citation2005), Rytioja et al. (Citation2014) and Malinovsky et al. (Citation2016). Notably, van den Brink & de Vries (Citation2011) provide schematic structures of specific plant celluloses, hemicelluloses and pectins that are very informative. In order to infect plants, fungi secrete CWDEs, breaching the cell walls through depolymerization of these polymeric components. Their actions facilitate entry into the host, while also releasing sugars that can be metabolized by the infecting fungus.
Investigation of fungal enzymes that can degrade plant cell wall components including cellulose, hemicellulose and lignin has been the focus of researchers searching for ways to degrade plant biomass or break down wood to provide pulp. These CWDEs or carbohydrate-active enzymes (CAZymes; Battaglia et al. Citation2011; van den Brink & de Vries Citation2011; Kubicek et al. Citation2014; Rytioja et al. Citation2014) are secreted by fungi in their natural environments to enable growth and reproduction. However, due to the occupation of different ecological niches, fungi vary in their carbon utilization patterns; enzymes that are secreted during infection vary greatly depending on the organism’s natural niche (Brown et al. Citation2014), so those secreted during natural growth and reproduction can differ from those secreted during the infection process. Broadly speaking, there are three major categories of CAZymes: (i) glycoside (glycosyl) hydrolases (GHs), which hydrolyse glycosidic bonds between oligo- or polysaccharides; (ii) carbohydrate esterases (CEs), which hydrolyse carbohydrate esters; and (iii) polysaccharide lyases (PLs), which cleave glycosidic linkages present in acidic polysaccharides (Lombard et al. Citation2014). Each CAZyme category consists of multiple families or groupings containing enzymes with specific activities, although differing families can also contain enzymes with similar functions (van den Brink & de Vries Citation2011). Detail about each CAZyme family involved in cellulose, hemicellulose and pectin degradation is the subject of many other major reviews, including one by Kubicek et al. (Citation2014). A focus here is to provide a review of the CWDEs/CAZymes present in plant pathogenic fungi including the model biotroph U. maydis.
Ustilago maydis CWDEs
Fungi secrete proteins to facilitate communication with their surroundings, as well as to ensure acquisition of nutrients. In plant pathogenic fungi, there are distinct differences in the profiles of secreted proteins depending upon their nutritional lifestyles. For example, of the secreted proteins identified in U. maydis, only approximately 25% are CWDEs, compared with about 50% in the basidiomycete Pycnoporus cinnabarinus and Phanerochaete chrysosporium white-rot fungi (Vanden Wymelenberg et al. Citation2006; Couturier et al. Citation2012; Levasseur et al. Citation2014). The number of CWDEs in biotrophs, including U. maydis, is also lower than in fungi with other modes of pathogenesis (Zhao et al. Citation2013), suggesting that reduced cell wall degradation activity is a characteristic of biotrophs to limit damage to the host and the subsequent elicitation of defence responses (Kämper et al. Citation2006; Mueller et al. Citation2008). Interestingly, a different view of CWDE production by U. maydis is presented when considering the possible biotechnological use of this fungus for biofuel production. Couturier et al. (Citation2012) indicated that U. maydis is a surprisingly good candidate for industrial use due to the high prevalence of proteins that hydrolyse hemicellulose and lignocelluloses into soluble sugar molecules. Additionally, Geiser et al. (Citation2016) illustrated the value of U. maydis for industrial biomass degradation through manipulation of the promoters of intrinsic CWDEs, including those that bioconvert xylans and celluloses into fermentable sugars. Furthermore, secretome analysis of a sequenced basidiomycete relative of U. maydis (Pseudozyma brasiliensis; Oliveira et al. Citation2014) revealed that this species contains enzyme sets capable of breaking down major polysaccharides in plants, such as xylan, indicating it is also a good candidate for the biomass degradation industry (Kaupert Neto et al. Citation2016). These reports seem to contradict the concept that U. maydis, as a biotroph, has lower cell wall-degrading enzyme activity. However, the studies may also indicate that U. maydis targets its release of suites of highly effective CWDEs during biotrophic development.
In U. maydis, sequencing the genome has enabled the identification of secreted proteins, including CWDEs (Kämper et al. Citation2006; Mueller et al. Citation2008). However, variation in the approaches used has resulted in the identification of slightly different lists of CWDEs. Kämper et al. (Citation2006) identified 168 putative secreted enzymes, 33 of which were CWDEs, and Doehlemann et al. (Citation2008a) provided expression data for 26 of these CWDEs. A subsequent investigation of the U. maydis secretome discovered 86 secreted proteins, 30 that were potentially involved in cell wall degradation (Couturier et al. Citation2012). Lanver et al. (Citation2014) detected 37 U. maydis proteins possibly involved in plant cell wall modification. More specifically, the CWDE repertoire of U. maydis includes cellulose-degrading β-endoglucanase and β-glucosidase enzymes, as well as a number of enzymes involved in processing xylan (Mueller et al. Citation2008; Battaglia et al. Citation2011; Rytioja et al. Citation2014). This is likely due to the large proportion of cellulose- and xylan-type molecules present in the maize cell wall (Carpita et al. Citation2001). Additionally, enzymes that target plant cuticle tissue and lignin were detected in the U. maydis secretome (Mueller et al. Citation2008). Ustilago maydis is not as well equipped with pectin-, xyloglucan- and galactomannan-associated enzymes (Battaglia et al. Citation2011), reflecting the limited presence of these types of molecules in maize tissue (Mueller et al. Citation2008) and the fact that pectin degradation is only required for apoplastic cavity development and not disease development throughout infection (Doehlemann et al. Citation2008a). This last point emphasizes the concept of targeted expression of potentially highly active enzymes. While genome sequencing reveals the presence of these enzymes, it does not indicate which ones are necessarily involved in pathogenesis.
To determine which CWDEs may be involved in U. maydis pathogenic development, we combined expression data from Kämper et al. (Citation2006), Doehlemann et al. (Citation2008a) and Skibbe et al. (Citation2010) to provide evidence of which cell-types the enzymes were transcribed in and whether the transcripts were present during in planta growth. These data are presented in and , while a comprehensive list of all identified U. maydis CWDEs is presented in Supplementary Table 1. It is noteworthy that the majority of the U. maydis CWDEs in this comprehensive list are members of the CAZyme category GH, along with some CEs, which is consistent with it being a biotroph. This group of enzymes imparts a diverse array of catalytic functions important for establishing infection by facilitating hyphal penetration of plant tissues through polysaccharide rearrangement in plant cell walls (Lanver et al. Citation2014; Rytioja et al. Citation2014).
Table 1. Up-regulated cell wall-degrading enzyme (CWDE) genes during pathogenic development of Ustilago maydis.
Table 2. Down-regulated cell wall-degrading enzyme (CWDE) genes during pathogenic development of Ustilago maydis.
Ustilago maydis penetrates cells of the host plant (maize) through the production of an appressorium, a specialized cell-type that facilitates breaching the cuticle layer and cell wall by releasing enzymes that depolymerize plant cell wall components. Sixteen potential U. maydis CWDEs, including a number of GHs, have differential transcript representation during appressorium and filament formation ( and ; Lanver et al. Citation2014) and corresponding lytic activity has been detected only at infection sites and not in the surrounding healthy plant tissue (Cano-Canchola et al. Citation2000). Notably, the expression of two U. maydis α-L-arabinofuranosidases, one GH51 and the other GH62 (UMAG_01829, afg1 and UMAG_04309, afg3, respectively), are up-regulated 12-fold and 4-fold in the appressorium, respectively. These enzymes act on arabinoxylan through depolymerization, releasing the constituent sugars arabinose and xylose. Interestingly, expression of afg1 was also over 1000 times up-regulated at 5 days post-infection (DPI), and continued to be highly expressed in each tumour stage assessed, suggesting that there is a requirement to degrade arabinoxylan throughout the pathogenic cycle (, Doehlemann et al. Citation2008a). This gene expression analysis further revealed increased transcript levels of CWDEs involved in cellulose and hemicellulose depolymerization during U. maydis pathogenesis, which occurred at 5 DPI and was maintained throughout pathogenesis (; Doehlemann et al. Citation2008a). Two endoxylanases, encoded by UMAG_04422 and UMAG_03411, were highly up-regulated at 5 DPI and 13 DPI, respectively (; Doehlemann et al. Citation2008a). Genes encoding enzymes for the degradation of cellulose and hemicellulose were up-regulated in a U. maydis–Arabidopsis thaliana infection model, where the fungus can grow, but not complete its sexual cycle (Méndez-Morán et al. Citation2005; Martínez-Soto et al. Citation2013). GH family enzymes, which contribute to the depolymerization of cellulose and arabinoxylan, were up-regulated during the initiation and progression of U. maydis infection.
Though there is evidence that CWDEs, including GHs, are critical for the establishment and progression of infection, the correlation between virulence and CWDEs is not always clear. It was previously suggested that an endoxylanase of U. maydis (UMAG_06350), an enzyme involved in the degradation of xylan, is not involved in U. maydis infection since its deletion resulted in a similar growth phenotype to the wild-type strain (Geiser et al. Citation2013). Similarly, deletion of the gene that encodes a GH45 family 1,4-β-endoglucanase (egl1) that is highly expressed during filamentous growth and tumour development did not affect filamentation or pathogenicity (Schauwecker et al. Citation1995). Even a triple knockout mutant, where three versions of egl were deleted, was no less virulent or pathogenic than the wild-type strain, and appressorium and filament formation was unaffected (Lanver et al. Citation2014). However, deletion of the α-L-arabinofuranosidases afg1, afg3 and afg2 (UMAG_00837) yielded a triple mutant strain with reduced virulence and host penetration efficiency, relative to wild-type strains (Lanver et al. Citation2014). The differences in reported findings is likely due to functional redundancy of CWDEs, where deletion of one or two genes may be complemented by genes encoding other similar enzymes. Coding an excess of CWDEs could mean that not all genes are required for pathogenesis. In this case, why would U. maydis contain multiple enzymes with redundant or overlapping functions? One explanation is to provide protection against mutation since, with this redundancy, DNA sequence changes in a single enzyme would not significantly reduce the ability of U. maydis to complete its pathogenic cycle due to the presence of other similarly functioning enzymes. This redundancy would ensure that ability to infect and acquire nutrients is protected in the fungus.
CWDEs for fungal carbon metabolism
Ustilago maydis, and plant pathogenic fungi in general, require CWDEs to facilitate infection by breaking down complex structural polysaccharides and releasing simple sugars. The products from this process could be consumed by the fungus, since they are biochemically indistinguishable from other sugar sources. As such, it is probable that biotrophic fungi synthesize and secrete CWDEs to facilitate penetration and growth within the plant, utilizing the sugars released, while also releasing effectors and phytohormones that act to redirect plant metabolism and make other carbon sources available.
Among the carbon sources used by fungi, glucose is preferred, with other hexose sugars such as fructose and mannose also being readily utilized. Pentoses, such as xylose and arabinose, can also be efficiently metabolized by fungi (Jeffries Citation1983; Seiboth & Metz Citation2011). Fungi are unable to directly metabolize large polysaccharides, and therefore must depolymerize these complex compounds to obtain sugars for uptake and subsequent metabolism. For example, while growing in culture, fungi can obtain glucose through amylase-mediated hydrolysis of starch, thus enabling them to use this polymer as a sole carbon source (Ohta Citation1997; Nahas & Waldemarin Citation2002). While growing in planta, the degradation of complex plant cell wall polysaccharides would result in the release of both pentose and hexose sugars that may then be taken up by fungi. However, sucrose and hexoses are also available from the redirection of plant metabolism. Fungal invertases and transporters are required to utilize these carbons (Talbot Citation2010).
Invertases
Invertases are responsible for converting intra- and extracellular sucrose into the monosaccharide components fructose and glucose, which are substrates for high-affinity sugar transporters. Invertases belong to multiple subclasses of glycoside hydrolases; GH32, GH43 and GH62 invertases have been identified in fungi (Lombard et al. Citation2014). They are further categorized based on their pH and mechanism of glycosidic bond cleavage (Van der Nest et al. Citation2015). The majority of enzymes in the GH32 family are functionally classified as acid invertases that can be cell wall-bound or vacuolar, and are involved in source-sink interactions and carbon processing in a number of phytopathogenic fungi (Pons et al. Citation1998; Lammens et al. Citation2009; Parrent et al. Citation2009). Fungi can acquire sugars such as inulin and sucrose, which are then depolymerized by acid invertases through α1-β2 glycosidic bond cleavage (Lammens et al. Citation2009). Secreted fungal invertases are released to the apoplast where they process sucrose transported, via the phloem, to the new sink at the site of fungal infection. The action of invertases increases the availability of free hexoses. The fungus takes up and utilizes the apoplast sugars (reviewed in Morkunas & Ratajczak Citation2014), maintaining the infection area as a sink. There are several examples of a correlation between plant infection and invertase activity. Infection by Puccinia hordei resulted in reduced sucrose levels in the apoplastic spaces of barley leaf tissues (Tetlow & Farrar Citation1992). Infection of wheat by P. graminis f. sp. tritici led to a slight increase in invertase activity, whereas white blister rust-infected A. thaliana induced a much greater change in its invertase protein AtβFruct1 (Heisterüber et al. Citation1994; Chou et al. Citation2000). Plasmodiophora brassicae, a biotrophic pathogen, causes clubroot on a number of vegetables and its infection is associated with increased invertase activity (Siemens et al. Citation2011). Activity from all invertases tested in powdery mildew infection of wheat leaves exhibited a marked increase compared with non-infected leaves (Sutton et al. Citation2007) and increased invertase transcription was observed in Aspergillus flavus-infected maize kernels (Dolezal et al. Citation2014). These plant-pathogen interaction studies provide evidence that increased invertase expression is linked to host plant infection. However, it is not always clear whether the activity is from plant- or fungal-derived invertases (Doidy et al. Citation2012). To help establish whether fungal invertase activity facilitates infection, use of a model system that can be manipulated at a molecular level, such as U. maydis, is required.
The U. maydis genome encodes two Suc2 invertases (UMAG_01945 and UMAG_03605), which are the only invertases characterized in this organism. Both are up-regulated in infected maize tissue (Kämper et al. Citation2006) and suc2 invertase activity increased as early as 2 DPI, suggesting that it is involved in the observed increase in free soluble sugars during infection (Horst et al. Citation2008). The lack of suc2 expression in haploid sporidia suggests it might be specific for filamentous growth in planta (Horst et al. Citation2008). The early induction (2 DPI) of suc2 during U. maydis infection is similar to that of the rust pathogen Uromyces fabae, in which the invertase uf-Inv1 was expressed during plant penetration, with high levels found in the haustoria (Voegele et al. Citation2006). In contrast, Voll et al. (Citation2011) detected increased invertase transcription only during later stages of infection of maize by U. maydis. Together, these studies suggest that U. maydis may be expressing invertases and in turn, stimulating the plant to produce and release invertases as well. They also suggest a changing requirement for invertase activity during fungal development within the plant. To determine if the invertase activity present at each stage of infection is of fungal or plant origin, U. maydis suc2-deficient strains and/or strains in which Suc2 proteins are tagged, could be created to assess the ability to establish infection without this enzyme and/or to follow the expression of the enzyme and the concomitant increase in free sugars during pathogenic development.
Transporters
Following the action of invertases and other enzymes, pathogenic fungi must take up the sugars made available. Transporters are responsible for facilitating entry of monosaccharide sugars into fungal cells. The diverse array of transporters present in some fungi contributes to their ability to process a variety of carbon sources and therefore grow in many different ecological environments. Sugar transporters are present at the plant-fungal interface, and are generally located in the host penetration structures, resulting in competition with the host plant for the pool of sugars released from the phloem into the apoplast (Voegele et al. Citation2001; Wippel et al. Citation2010). Fungal sugar transporters belong to the major facilitator superfamily and are predicted to have a similar structure, composed of 12 transmembrane domains with a large extracellular loop between the first and second domain (Wahl et al. Citation2010; Doidy et al. Citation2012). The predicted conserved structure of this superfamily of proteins suggests that they execute similar functions – that is, the uptake of sugars into cells.
Functional conservation exists among fungal hexose transporters but sugar preference of transporters in specific species may vary. For example, U. fabae Hxt1 preferentially takes up glucose and fructose (Voegele et al. Citation2001), while the monosaccharide transporter Frt1 in Botrytis cinerea, a necrotrophic fungus causing grey mould in grapes, has a high affinity for fructose (Doehlemann et al. Citation2005). The monosaccharide transporters of mycorrhizal fungi can take up pentoses released from cell wall depolymerization, such as xylose (Helber et al. Citation2011). In the Globus sp. system, increased xylose induces expression of monosaccharide transporters in the extraradical mycelium (Helber et al. Citation2011). In the hemibiotrophic maize pathogen Colletotrichum graminicola, four of the five characterized hexose transporter genes are differentially regulated depending on the mode of nutrition, likely reflecting different sources of carbon available during the biotrophic and necrotrophic lifestyle. CgHxt1, CgHxt2 and CgHxt3 are up-regulated during the biotrophic phase and can accept a number of hexose sugar substrates, but have a lower preference for pentoses such as xylose (Lingner et al. Citation2011). The 19 members of the sugar transporter family that have been identified in U. maydis are listed in (Kämper et al. Citation2006; Wahl et al. Citation2010). The most well-characterized hexose transporter is Hxt1 (UMAG_05023; Schuler et al. Citation2015), which has been shown to transport glucose, fructose and mannose with very high affinity (Schuler et al. Citation2015). In addition, when the U. maydis Hxt1 was expressed in a monosaccharide transporter-deficient Saccharomyces cerevisiae strain, it was shown to have a low affinity for xylose (Schuler et al. Citation2015). Sugar preference in hexose transporter uptake may reflect sugars present in a given host or at a given stage of disease development. In this regard, it is interesting that U. maydis also codes a sucrose transporter, Srt1 (UMAG_02374), so it has the capacity to take up sucrose as well as monosaccharide sugars (Wahl et al. Citation2010).
Table 3. Pathogenesis and microarray expression data of Ustilago maydis sugar transporter family members.
Srt1 is the only U. maydis transporter characterized that can directly transport sucrose into cells (Wahl et al. Citation2010). By taking up sucrose directly from the apoplast, the fungus may avoid the initiation of plant defence responses shown to be related to the uptake of glucose (Herbers et al. Citation1996; Rolland et al. Citation2006; Wahl et al. Citation2010). Ustilago maydis Srt1 also has a much higher affinity for sucrose compared with the maize sucrose transporter Sut1. This suggests that U. maydis could outcompete its host for the uptake of sucrose from the apoplast, bypassing the need for invertase activity and hexose uptake which may, in turn, provide a special advantage to its biotrophic lifestyle.
No matter which pathogenic lifestyle a fungus has adopted, it seems logical that sugar transporters would be required for pathogenic growth, and expression data support this. Erysiphe cichoracearum infection of A. thaliana as well as Erysiphe necator- and Plasmopara viticola-infection of grapevine leaves, result in up-regulation of their respective sugar transporter proteins (Fotopoulos et al. Citation2003; Sutton et al. Citation2007; Hayes et al. Citation2010). Similarly, hexose transporter proteins from Fusarium graminearum (Kruger et al. Citation2002) and Blumeria graminis (Zhang et al. Citation2005) are expressed during plant infection. Microarray analysis of 16 U. maydis transporter genes showed that 12 genes are up-regulated in 13 DPI tumours compared with growth in axenic culture (; Kämper et al. Citation2006). The expression of srt1 is induced by in planta growth, peaking when tumours form, and is not present in fungal cells grown in axenic culture with added sucrose or glucose, indicating that this transporter requires signals from within the plant to be activated, and it is not subject to carbon catabolite repression (Wahl et al. Citation2010). In contrast, when U. maydis is inoculated on the non-host A. thaliana, srt1 is down-regulated early during infection (Martínez-Soto et al. Citation2013). This species-specific expression suggests that its direct function is to take up sucrose at the plant-fungal interface only during colonization of its natural host, maize. The common outcomes of expression analysis in a number of plant pathogens suggests that there is a broad conservation of transport function, especially among proteins related to Hxt1. However, many fungi also have multiple transporters, so questions remain about which one(s) are required for pathogenic growth.
Ustilago maydis deletion analyses revealed that the sucrose transporter Srt1, and the hexose transporter Hxt1, are each required for full pathogenic development and disease symptom formation (Wahl et al. Citation2010; Schuler et al. Citation2015). Even though the srt1 deletion mutants show dramatic reduction in pathogenicity, they retain the ability to cause disease, and this ability is reduced further in the double deletion of srt1 and hxt1 (Schuler et al. Citation2015). Pathogenesis was restored when the sucrose transporter suc9 from Arabidopsis was expressed from the srt1 promoter in the ∆srt1 mutant strain (Sauer et al. Citation2004; Wahl et al. Citation2010). However, when Hxt1 function was complemented with hexose transporters CgHxt3 and Stp1 from A. thaliana, pathogenesis was still reduced, which the authors attribute to the presence of a sensor capability in addition to the transporter function of Hxt1. When the region of the protein predicted to provide this sensor capability was mutated, making it a constitutively active sensor, the resulting U. maydis strain was completely non-pathogenic (Schuler et al. Citation2015). Separate investigations revealed that Srt1 and Hxt1 are present in the plasma membrane of hyphae, and similarly, the sugar transporter UfHxt1, which shows 50% identity to the U. maydis Hxt1, is localized to the haustoria plasma membrane of U. fabae, supporting a role for these transporters in the establishment of infection (Voegele et al. Citation2001; Wahl et al. Citation2010; Schuler et al. Citation2015). Together, the results are consistent with Srt1 having a role in sucrose uptake during pathogenesis, while the role of Hxt1 as a sensor is more prominent than its role as a hexose transporter. Redundancy may explain this observation, as noted earlier in regards to CWDEs; the presence of multiple proteins with hexose transporter function may provide redundancy for the Hxt1 transporter activity, but not for its sensing ability. Thus, it is likely that many of the sugar transporters in U. maydis contribute to pathogenic development, and the production of a variety of transporters may be a requirement because the types of sugars available for uptake may vary during pathogenic development and/or depending on the plant tissue infected. This variation would further suggest differential regulation of the transporters to ensure adequate carbon uptake throughout pathogenic development.
Carbon catabolite repression (CCR)
Although a number of pentose or hexose sugar sources are available to fungi in plant hosts, and fungi can alter which carbon catabolism pathways are activated (Ruijter & Visser Citation1997), they prefer carbon sources that are readily available and easily metabolized (Kelly Citation2004). Carbon catabolite repression is a regulatory mechanism to increase the utilization of favourable carbon sources by ensuring the repression of synthesis of enzymes required to metabolize alternative or secondary carbohydrate sources (Ruijter & Visser Citation1997; Gancedo Citation1998). In cases where multiple sugar types are available, one is typically preferred over others; glucose provides the best energy gain and is therefore the most favourable carbon source for fungal cells. In instances where glucose is available, the expression of other catabolite pathways is often suppressed (Brown et al. Citation2014). This regulation is beneficial because it limits utilization of energetically unfavourable carbon compounds and suppresses expression of carbohydrate metabolism genes that are not required. Mutations in CCR regulatory mechanisms have shown that relief from CCR is adequate for utilization of non-preferred carbon sources (Chang & Todd Citation2004). Investigating CCR mechanisms in plant pathogenic fungi is important for understanding how these pathogens thrive throughout disease development; yet the vast majority of information on this process has come from studies of ascomycete fungi in culture. An overview of the findings from these studies is presented below to provide context for discussions of plant pathogens and basidiomycete fungi.
Ascomycetes
The term ‘carbon catabolite repression’ is often interchangeably referred to as ‘glucose repression’, since glucose frequently represses the expression of genes involved in processing other sugars. In the filamentous ascomycete Aspergillus nidulans, a number of enzymes involved in the degradation of plant carbons, such as xylan, cellulose and pectin are repressed by glucose; however, other sugars may also initiate CCR (Ruijter & Visser Citation1997; de Vries & Visser Citation2001). CCR induction by other sugars has also been noted in a strain of S. cerevisiae that was engineered to metabolize xylose. In this strain, the presence of xylose represses the expression of sucrose- and galactose-metabolizing genes (Belinchón & Gancedo Citation2003). CCR is controlled in ascomycetes by a transcription factor that is differentially phosphorylated depending upon whether glucose is present. When glucose is available, genes for metabolizing other carbon sources (glucose repressible genes) are turned off, but are turned on when glucose is present at low levels or is absent. In Aspergillus species, the controlling transcription factor is CreA, which was recently identified as an essential growth factor; a conserved region of CreA in Aspergillus species and Trichoderma reesei is required for growth on numerous carbon, nitrogen and lipid sources (Ries et al. Citation2016). In creA deletion studies, a number of cell wall-targeting genes were induced, indicating that they are directly repressed by CreA in the presence of secondary carbon sources such as galactose or xylose (Culleton et al. Citation2013), once again indicating that glucose is not the only sugar capable of initiating CCR and linking CWDEs to CCR.
The control of CCR has been extensively studied in the ascomycete genera Saccharomyces and Aspergillus (Ronne Citation1995; Ruijter & Visser Citation1997; Brown et al. Citation2014). In S. cerevisiae, the DNA-binding protein Mig1 is the key glucose repression regulator (reviewed in Gancedo Citation1998). When glucose is available, Mig1 is shuttled into the nucleus (DeVit et al. Citation1997) and represses transcription of glucose-repressible genes by binding to their promoters and recruiting a Ssn6 (Cyc8)-Tup1 co-repressor complex (Nehlin et al. Citation1991; Papamichos-Chronakis et al. Citation2004). In response to glucose limitation, a serine/threonine kinase, sucrose non-fermenting 1 protein (Snf1) regulates phosphorylation of Mig1 (Wilson et al. Citation1996; Ostling & Ronne Citation1998). When Mig1 is phosphorylated, it is exported to the cytoplasm (DeVit et al. Citation1997), making interaction with the Ssn6 (Cyc8)-Tup1 complex no longer possible and resulting in transcription of glucose-repressible genes (Papamichos-Chronakis et al. Citation2004). Mig1-deficient strains show loss of CCR and have impaired growth (Dowzer & Kelly Citation1991). In filamentous ascomycetes, such as A. nidulans, the transcription factor CreA/CRE1 is a functional homologue of the Mig1 in S. cerevisiae (Ostling et al. Citation1996; Strauss et al. Citation1999). In the presence of glucose, CreA/CRE1 binds to upstream regulatory elements, blocking transcription of enzymes required to process alternate carbon sources (Brown et al. Citation2014). In carbon-limiting environments, CreA/CRE1 is phosphorylated by SnfA, an orthologue to Snf1, and it is exported from the nucleus, allowing induction of carbon metabolism genes (Brown et al. Citation2014). Similarly to ∆mig1 mutants, creA-null strains exhibit impaired growth and show loss of CCR in the presence of glucose (Dowzer & Kelly Citation1991). For further information on these transcriptional regulation mechanisms, see reviews by Aro et al. (Citation2005), Brown et al. (Citation2014) and Fernandez et al. (Citation2014). This well-characterized control of CCR and the link to repressing CWDEs make it a reasonable candidate for control of carbon metabolism during pathogenesis by plant pathogenic fungi.
The initiation of CCR is stimulated by Snf1/SnfA, whose activation results in the derepression of alternative carbon source-related genes including CWDEs. CWDEs have been identified as pathogenesis factors (reviewed in Kubicek et al. Citation2014), suggesting that the CCR pathways would contribute to pathogenesis if CWDEs were regulated by CCR activator or repressor proteins. As such, functional investigations of the Snf1 kinase have been carried out in some ascomycete plant pathogens. For example, in Cochliobolus carbonum, F. oxysporum and Leptosphaeria maculans, deletion of snf1 resulted in repression of many cell wall-targeting lytic enzymes and reduced virulence (Tonukari et al. Citation2000; Ospina-Giraldo et al. Citation2003; Feng et al. Citation2014). Deletion of the Magnaporthe oryzae snf1 homologue did not have the same transcriptional effect on CWDEs, but resulted in impaired appressorium formation and pathogenicity (Yi et al. Citation2008). Since Snf1 regulates glucose-repressed genes, cells that lack this kinase are unable to utilize carbon sources other than glucose, possibly explaining the deletion pathogenesis phenotypes in some fungi. To date, only a single creA deletion mutant has been functionally assessed in ascomycete phytopathogens. Deletion of the creA and snf1 homologs in Alternaria brassicicola did not affect pathogenesis and did not influence expression of CWDEs, suggesting that these CCR proteins are not involved in regulating carbon uptake or pathogenesis in this fungus (Cho et al. Citation2009). Together, the data indicate that for some, but not all ascomycete plant pathogens, CCR plays a role in controlling the expression of CWDEs and carbon uptake during pathogenic development.
Basidiomycetes
While most of the carbon catabolite research has been conducted on ascomycetes, recent genome sequence information provides insight into possible CCR mechanisms in the basidiomycetes. As in ascomycetes, CreA homologues in basidiomycetes could mediate repression of carbon metabolism or CWDEs in the presence of various carbon sources (Rytioja et al. Citation2014). A comparative genomics study identified CreA homologues in 27 of the 31 basidiomycete genomes assessed. These included U. maydis (UMAG_04909) and the white rot fungus Phanerochaete chrysosporium (Todd et al. Citation2014). Mig1 homologues were not detected in any of the basidiomycetes. However, the fact that most basidiomycete species contain a CreA homologue suggests its conserved role as a transcriptional regulator of carbon metabolism in these fungi, though there have been no functional studies to this effect in the basidiomycetes. The function of CCR in relation to the control of CWDE expression has been investigated in a limited number of basidiomycete fungi. Early studies, conducted before genome sequence data were available, included only a single phytopathogen; Athelia rolfsii (Sclerotium rolfsii) (Shewale & Sadana Citation1981; Chow et al. Citation1994; Onishi & Tanaka Citation1996; Ding et al. Citation2001). More recent investigations of plant pathogenic basidiomycetes reveal that CCR mechanisms have a role in CWDE expression in the common corn smut pathogen and white-rot fungi. In U. maydis, Snf1 (UMAG_11293) negatively regulates the expression of two xylanases and positively regulates an endoglucanase and a polygalacturonase. Deletion of snf1 resulted in a slight reduction in pathogenic development; however, it is not required for releasing the fungus from glucose repression since ∆snf1 mutants can grow on alternative carbon sources in axenic culture (Nadal et al. Citation2010). Therefore, in contrast to the situation in ascomycetes, U. maydis Snf1 positively and negatively regulates some CWDEs, but is not a global glucose regulator (Nadal et al. Citation2010). However, this distinction between the ascomycetes and basidiomycetes is not universal, as white rot basidiomycete fungi have a CCR-mediated mechanism for suppressing CWDE expression. In the presence of a glucose source, cellulase (endoglucanase) and xylanase are repressed, and when glucose is depleted, transcription of these enzymes resumes (Kobakhidze et al. Citation2016). These limited studies provide some insight regarding the link between CCR and CWDE expression and pathogenesis in basidiomycetes; however, it is clear, especially for U. maydis, that the transcriptional regulation of CWDEs by glucose repression regulatory machinery and the resulting impact on pathogenesis is influenced by signalling mechanisms directed through Snf1 as well as other proteins. Furthermore, the expression of CWDEs does not occur with the complete suppression of other sugar metabolizing enzymes (). The nature of CWDE gene expression control and the shifts in expression in U. maydis require further investigation.
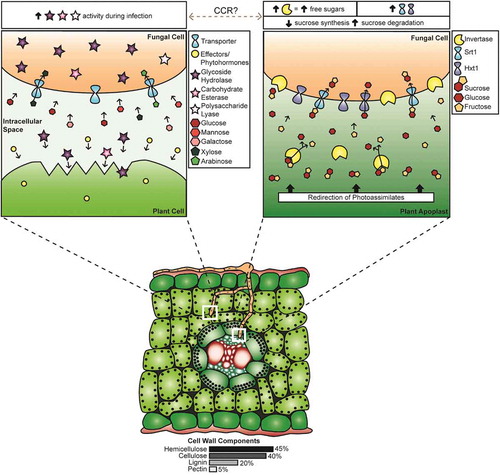
Fig. 1 (Colour online) Diagrammatic representation of events related to carbon acquisition by Ustilago maydis during pathogenic growth in maize (Zea mays). (Bottom panel) Cross-section of a maize leaf, showing that fungal mycelia have penetrated the cuticle and epidermis, growing between and through the mesophyll cells. The upper mycelium is in the process of penetrating the next mesophyll cell while the lower mycelium has penetrated the bundle sheath and has grown between the phloem cells. The white boxes indicate the areas with expanded views. (Upper left panel) Expanded view of mycelium approaching a plant mesophyll cell. Ustilago maydis modifies the host physiology and suppresses defence responses through the action of effectors and phytohormones, while CAZymes act on the cell wall. The approximate proportions, based on the comprehensive U. maydis CAZyme list in Supplementary Table 1, of GH (~76%), CE (~23%) and PL (<1%) enzymes are indicated by the number of coloured stars. These enzymes depolymerize cell wall components, releasing hexose and pentose sugars which are taken up by the fungus via transporters; the proportion of cell wall components are indicated below the lower panel. The text box above this panel indicates increased CWDE activity during infection. (Upper right panel) Expanded view of carbon uptake by a mycelium interacting with the phloem tissue, representing the results of U. maydis creating sink tissue in the leaves. Sucrose moves through the phloem and is available in the leaf vasculature for direct uptake by the fungus via Srt1. Alternatively, sucrose is acted upon by cell wall or vacuolar invertases to yield glucose and fructose, which are transported into the fungus via the monosaccharide sugar transporter Hxt1. The text box above this panel indicates the role of CCR in this process; increased invertase and transporter activity, increase in free sucroses, decrease in sucrose synthesis genes, and an increase in sucrose degradation genes is observed in infected tissue. The switch between methods of carbon acquisition by the fungus, which is indicated by the grey dashed line, could be mediated by CCR systems.
Plant defences and biotrophic interactions
Biotrophic fungal pathogens obtain nutrients from living plant tissue; however, to access these nutrients, they must overcome a barrage of plant defences. Plants have physical barriers such as a cuticle and rigid cell walls, as well as signal response systems that can lead to the reinforcement of physical barriers through callose or lignin deposition, the expression of defence proteins, or the synthesis of small molecules including the creation of reactive oxygen species (reviewed in Zeilinger et al. Citation2016). They may even sacrifice cells through the hypersensitive response to ward off infection (Heath Citation1998). These defences are implemented at different stages of infection and are not static. Plants constantly monitor for the presence of microorganisms through the receptor-like kinases, which act as pattern recognition receptors detecting the presence of microbes, pathogens or damage-associated molecular patterns (MAMPS, PAMPs or DAMPs) (Kushalappa et al. Citation2016; Eckardt Citation2017). The damage that may be detected includes changes to cell wall structure through the action of pathogen-secreted CWDEs (reviewed in Malinovsky et al. Citation2016). Upon plant detection of one of these patterns, there is a cascade response leading to the activation of defence mechanisms and plant immunity that has been referred to as pattern- or PAMP-triggered immunity (PTI). However, pathogens, including fungal biotrophs, produce and release protein effectors and hormones that can alter host physiology and circumvent this immunity (Bakkeren & Valent Citation2014; Morrison et al. Citation2015; Matei & Doehlemann Citation2016). The battle continues with the plant, which in turn responds with effector-trigger immunity or ETI, another cascade defence response. With this massive capacity for defence, it would seem an insurmountable task for fungal biotrophs to access nutrients from living plant tissue, yet they do. One approach to biotrophy is to colonize the plant without triggering the defense responses mentioned above. This involves altering plant responses and physiology through the release of effectors and phytohormones, as well as controlling how and when carbon sources are accessed.
Ustilago maydis initiates its biotrophic infection of maize by altering gene expression in response to sensing plant surface cues, such as the hydrophobicity or the presence of cutin monomers (Lanver et al. Citation2014). As mentioned earlier, this alteration includes the expression of the arabinofuranosidases GH51 and GH62, which are active during plant penetration (Lanver et al. Citation2014; ). Effectors that suppress plant defences are also expressed and secreted during the early stages of infection. Functions have been identified for several effectors; Pep1 aids in suppressing the maize oxidative burst by inhibiting its POX12 peroxidase (Hemetsberger et al. Citation2012, Citation2015), while Pit2 suppresses the activity of cysteine proteases (Mueller et al. Citation2013). Pep1 and Pit2 suppress plant defences in the apoplast early during infection, while effector Cmu1 regulates chorismate homeostasis, resulting in the suppression of salicylic acid synthesis (Djamei et al. Citation2011), and Tin2 stabilizes a maize kinase which stimulates anthocyanin biosynthesis by redirecting precursors that would be used for lignin biosynthesis (Tanaka et al. Citation2014). This suppression of plant defences allows U. maydis to penetrate and begin colonizing maize tissues.
Table 4. Summary of Ustilago maydis infection of Zea mays leaves at various stages of growth.
We propose that, during this early phase of leaf infection, U. maydis acquires a portion of its carbon through the depolymerization of cell wall components ( upper left panel; ). As the infection progresses, U. maydis mycelia reach the leaf vascular bundles and carbon acquisition is augmented by, or may switch to, the uptake of sucrose and hexoses, the latter being released by the action of invertases ( upper right panel; ). At this stage of infection, the fungus also stimulates the expansion of mesophyll cells (hypertrophy) and the division of bundle sheath cells (hyperplasia), likely via the release of effectors and hormones. These alterations in cell morphology are a component of influencing the plant source-sink relationships, which are further altered by the synthesis of cytokinins and the alteration of cytokinin metabolism in maize by U. maydis (Bruce et al. Citation2011; Morrison et al. Citation2015, Citation2017). In an uninfected plant, source tissues, such as mature leaves, provide excess photosynthates to sink tissues such as developing leaves (Biemelt and Sonnewald Citation2006). The relative strength of each sink tissue within a plant determines the distribution of photoassimilates (Lemoine et al. Citation2013). Ustilago maydis-colonized maize tissue remains a strong sink throughout the infection cycle (Horst et al. Citation2008). Horst et al. (Citation2010) proposed that soluble sugars acquired through the redirection of nutrients from source tissues provide the fungus with a source of food and energy. This suggestion is supported by the work presented here ( upper right panel). Furthermore, we propose that the requirement for carbon acquisition by the biotrophic fungal pathogens provides a major selective pressure for effector evolution.
The influence of U. maydis on the host to increase sugar availability must be controlled. In feeding experiments where sugars were added to the sites of U. maydis infection, Kretschmer et al. (Citation2016) observed an increase in disease rating in their seedling assays and suggested that this would lead to increased plant responses, including turning on of defence processes. The plant response to infection was assessed by transcriptome analyses of U. maydis-infected developing maize leaves. It was noted that expression of photosynthesis-related genes, as well as those involved in sucrose and starch synthesis, were not activated as they would be in an uninfected leaf. As a result, the infected leaf remains a sink and does not become a photosynthetically active source (Doehlemann et al. Citation2008b; Kretschmer et al. Citation2016; Cheung et al. CitationForthcoming 2017). Infected tissues also exhibited an overall increase in soluble sucrose and hexoses, the latter being attributed to increased invertase activity (Horst et al. Citation2008; Doehlemann et al. Citation2008b). The source of the invertases – fungal or plant – is difficult to discern (Lemoine et al. Citation2013). However, altering sucrose concentration through the action of plant invertases and altered plant sucrose transporter expression leads to induction of pathogenesis-related gene expression (Tauzin and Giardina Citation2014). Thus, if plant invertases are involved, there must be some counteraction from U. maydis to ensure defence responses remain suppressed. The increased expression of the U. maydis Srt1, which has a higher affinity for sucrose than the maize sucrose transporters (Wahl et al. Citation2010), could contribute to this continual suppression of plant defences. It may also be that plant invertases have a limited role in increasing hexose concentration, since fungal invertases are up-regulated during infection. However, the molecular basis of the controlled increase in sugar availability by U. maydis requires further investigation.
To ensure carbon sources are available for the completion of its pathogenic/sexual cycle, U. maydis requires the ability to access different carbon sources, depending upon the stage of fungal development and the progression of disease. The transition in carbon acquisition between the stages of penetration and growth in the leaf mesophyll relative to acquisition once U. maydis reaches the vasculature and stimulates cell morphology changes has been discussed (; ). Recent work with effector proteins suggests there could be another switch in carbon sources during teliosporogenesis. Tollot et al. (Citation2016) showed that regulation of teliospore formation included repression of effector genes that were expressed during the early stages of U. maydis infection. Down-regulation of ‘early’ effectors during the formation of teliospores suggests that there is no longer a requirement to extract nutrients from the host without eliciting a defence response. At the stage when teliospore development begins, the fungus has already colonized the plant and established mechanisms of gaining nutrients that involve the expression of effectors, but now it must transition to fuelling the development of dispersal spores with stored carbon sources. There is very limited research into fungal carbon acquisition during sporogenesis. Cano-Canchola et al. (Citation2000) noted a correlation between a polygalacturonase (PG) as well as other PL family enzyme activity and teliospore formation. In addition, Castruita-Domínguez et al. (Citation2014) discovered that PG is involved in teliospore formation and tumour development of U. maydis. We propose that during sporogenesis, increased activity of PG and other PL family members is associated with increased pectin degradation. Breaking down pectin, which helps to hold cells together, would make the plant cells friable, while also providing space for the production of teliospores within tumours (). This proposed breakdown of pectin would also result in the release of α-1,4-polygalacturonic acid, which could be taken up by U. maydis via its galacturonic acid transporter, UMAG_00061, shown to be a potential orthologue of Neurospora crassa (Benz et al. Citation2014). We propose that, together, these observations illustrate several transitions in sugar acquisition by U. maydis during its development in planta. Further, we propose that these transitions could be influenced by CCR systems, suggesting a role for CCR in modulating the relative levels of cell wall degradation and free sugar uptake during the pathogenic cycle.
Conclusions and future work
A key focus of this review is the identification of shifts in carbon sources accessed by biotrophic fungi from plants during the infection process. Initially, they obtain carbon compounds via depolymerization of plant cell wall material, then the uptake of free sucrose and hexoses becomes a significant component of carbon metabolism, followed by a shift to pectin degradation and uptake of galacturonic acid. Since these shifts include transitions away from and back to complex carbohydrate degradation, CCR may be involved. We point out that CCR mediates the shift in carbon metabolism in ascomycete fungi, but that CCR does not control the switch from glucose metabolism to the expression of CWDEs to the same degree in basidiomycete fungal pathogens, possibly because there is a requirement for these fungi to access several carbon sources simultaneously. Thus, while the main carbon source may shift, this is not an ‘all-or-none’ response, and the fungus retains uptake and metabolism of carbons from other sources. Nonetheless, these shifts are prominent and, at least temporally, linked to shifts in disease progression, fungal development and effector gene expression.
It is possible that sensing carbon sources is a component of the control of developmental shifts in U. maydis during pathogenesis. Glucose sensing controls morphological shifts in the human pathogen Candida albicans (Miwa et al. Citation2004). Similarly, nitrogen sensing via ammonium permease has a role in the shift between the non-pathogenic budding haploid cells and the pathogenic filamentous dikaryon in U. maydis (Smith et al. Citation2003; reviewed in Braunsdorf et al. Citation2016). Thus, sensing nutrients, and specifically carbon, might be expected to influence development during fungal biotrophic growth. If this is the case, it would also provide a means of sensing the physiological state of the host, possibly linking the timing of fungal development to plant development, a key component of fungal pathogenesis in U. maydis. Further investigations into the control of carbon metabolism shifts by U. maydis would provide insight into the regulation of biotrophic fungal pathogenesis.
Carbon metabolism transitions are also linked to shifts in effector gene expression. It is possible that a common control mechanism exists, or that carbon sensing directs a shift in carbon metabolism and in effector expression to facilitate access to new carbon sources. To further investigate these possible linkages, some of the infection time-course RNA-seq and microarray datasets could be re-evaluated. However, a current limitation to this type of transcriptome analysis is that the existing data may not include important time points, especially those spanning the transition to teliospore formation. Therefore, a specific investigation of this important time period by RNA-seq is warranted. Another investigative approach would involve determining if varying carbon sources alter effector gene expression.
The shifts in carbon source acquisition during fungal pathogenesis must be considered when investigating whether a carbon metabolism or CWDE gene is required for successful infection of a potential host plant. If there is a shift, but it is not an ‘all-or-none’ shift, deletion of a gene encoding either of these functions would not be expected to block pathogenesis, but rather it may inhibit normal disease development in an incremental manner. Further, one might expect that timing of expression of carbon metabolism enzymes or CWDEs may be critical for successful growth within the plant. In this regard, recent findings that deletion of a xylitol dehydrogenase gene reduces pathogenesis in U. maydis are relevant (Goulet et al. unpublished data). As noted earlier, deletion of sucrose- or hexose-metabolizing enzymes reduces pathogenesis; however, the impact of blocking xylose metabolism had not been investigated. The depolymerization of maize cell wall polymers releases xylose molecules, suggesting xylose-containing polymers are a readily available and important carbon source within the maize plant. Interestingly, Mondo et al. (personal communication) have determined through comparative genomic analyses that the smut fungi have specific set CWDEs that target xylose-containing molecules, and these sets differ from other groups of basidiomycete fungi. Along with this, the xylitol dehydrogenase deletion pathogenesis phenotype is not completely complemented by constitutive expression of the gene, suggesting that timing of its expression may be critical to its full impact on pathogenesis. Together, these results suggest a role for xylose metabolism in pathogenesis by U. maydis and other smut fungi. It is also possible that the metabolism of other specific carbon sources is required for the full expression of pathogenesis. In this context, the shift providing access to galacturonic acid and its potential link to teliospore development should be investigated.
Determining the roles of CWDEs during pathogenesis should also take into account the possibility of having a required time-period of expression. Initially, the expression profile of CWDEs could be determined over a time course of pathogenesis to confirm differential expression at specific stages in disease development, followed by deletion analysis. Given the redundancy in CWDE functions, determining their roles in pathogenesis might require the deletion of multiple genes. The recently developed CRISPR-mediated genome modification methods for U. maydis could facilitate such a functional investigation (Schuster et al. Citation2016). Further, it is possible to create a U. maydis strain containing the minimum number of required CWDEs as a starting point for dissecting the roles of cell wall degradation during pathogenesis and the link to carbon uptake.
During in planta growth, U. maydis cells undergo developmental transitions and it is possible that these are linked to changes in carbon source availability. For example, many fungi enter into meiosis in response to nutrient limitation (reviewed in Saville et al. Citation2012), so investigating the impact of carbon source shifts, including the uptake of galacturonic acid, on the initiation of teliospore production could be informative. One approach may be to determine the impact of growing U. maydis on single carbon sources such as galacturonic acid or specific sugars, on the expression of transcriptional regulators of meiosis like unh1 (Doyle et al. Citation2016). A more global approach could be to carry out a metabolomics analysis over a time course of pathogenic development and noting the shifts in sugar or sugar acid presence relative to fungal developmental shifts. Such a study would capture sugars from the breakdown of cell walls and the changes in sugar availability throughout the maize growing season, assessing proposed shifts in carbon sequestration as outlined in this review. These metabolomics studies could be carried out in conjunction with transcriptomic studies to link the expression profile of specific genes to shifts in carbon source availability.
In considering transcriptome studies, it is also important to pay closer attention to subtle changes in transcription of carbon metabolism or CWDE genes, since the evidence presented in this review suggests that there is not likely to be a single carbon acquisition strategy that is responsible for fungal biotrophic pathogenic development. Since the ability of U. maydis to cause disease appears to require subtle changes in multiple carbon-related genes, elucidating what the subtle changes are and how they are controlled will be critical in developing a full understanding of carbon metabolism during fungal biotrophic growth. In general, the information presented herein indicates that the links between carbon metabolism and pathogenesis are complex and there is a need to focus on subtle changes in pathogenesis assays when investigating these relationships. Understanding carbon relations in fungal biotrophs is at an early stage; however, new tools that can be applied to model systems like U. maydis, and the ability to link findings to less tractable fungi through comparative transcriptomics, offers great promise for a rapid expansion of knowledge in this area. The key finding that biotrophs shift carbon source accessibility throughout the pathogenic cycle may be beneficial in directing future research in basidiomycete biotrophic pathogenesis.
Supplementary Table 1. Comprehensive list of potential carbohydrate-active/cell wall-degrading enzymes in U. maydis and their involvement in pathogenesis.
Download PDF (71.4 KB)Acknowledgements
We would like to acknowledge Ibraheem Alimi, Dr Colleen Doyle and Dr Erin Morrison for critical review of this manuscript. We would also like to thank Lauren Ostrowski (University of Toronto, Department of Laboratory Medicine and Pathobiology) for creation of the figure. Funding for this project was awarded through the Natural Sciences and Engineering Research Council (NSERC) of Canada (BJS, KMG) and the Ontario Graduate Scholarship (KMG).
Supplemental material
Supplemental data for this article can be accessed online here: https://doi.org/10.1080/07060661.2017.1354330.
References
- Aro N, Pakula T, Penttilä M. 2005. Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev. 29:719–739.
- Bakkeren G, Valent B. 2014. Do pathogen effectors play peek-a-boo? Front Plant Sci. 5:731.
- Banuett F, Herskowitz I. 1996. Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis. Development. 122:2965–2976.
- Battaglia E, Benoit I, van den Brink J, Wiebenga A, Coutinho PM, Henrissat B, de Vries RP. 2011. Carbohydrate-active enzymes from the zygomycete fungus Rhizopus oryzae: a highly specialized approach to carbohydrate degradation depicted at genome level. BMC Genom. 12:38.
- Belinchón MM, Gancedo JM. 2003. Xylose and some non-sugar carbon sources cause catabolite repression in Saccharomyces cerevisiae. Arch Microbiol. 180:293–297.
- Benatti MR, Penning BW, Carpita NC, McCann MC. 2012. We are good to grow: dynamic integration of cell wall architecture with the machinery of growth. Front Plant Sci. 3:187.
- Benz JP, Protzko RJ, Andrich JMS, Bauer S, Dueber JE, Somerville CR. 2014. Identification and characterization of a galacturonic acid transporter from Neurospora crassa and its application for Saccharomyces cerevisiae fermentation processes. Biotechnol Biofuels. 7:20.
- Biemelt S, Sonnewald U. 2006. Plant-microbe interactions to probe regulation of plant carbon metabolism. J Plant Physiol. 163:307–318.
- Braunsdorf C, Mailänder-Sánchez D, Schaller M. 2016. Fungal sensing of host environment. Cell Microbiol. 18:1188–1200.
- Brown NA, Ries LN, Goldman GH. 2014. How nutritional status signalling coordinates metabolism and lignocellulolytic enzyme secretion. Fungal Genet Biol. 72:48–63.
- Bruce SA, Saville BJ, Emery RJN. 2011. Ustilago maydis produces cytokinins and abscisic acid for potential regulation of tumor formation in maize. J Plant Growth Regul. 30:51–63.
- Cano-Canchola C, Acevedo L, Ponce-Noyola P, Flores-Martínez A, Flores-Carreón A, Leal-Morales CA. 2000. Induction of lytic enzymes by the interaction of Ustilago maydis with Zea mays tissues. Fungal Genet Biol. 29:145–151.
- Carpita NC, Defernez M, Findlay K, Wells B, Shoue DA, Catchpole G, Wilson RH, McCann MC. 2001. Cell wall architecture of the elongating maize coleoptile. Plant Physiol. 127:551–565.
- Castruita-Domínguez JP, González-Hernández SE, Polaina J, Flores-Villavicencio LL, Alvarez-Vargas A, Flores-Martínez A, Ponce-Noyola P, Leal-Morales CA. 2014. Analysis of a polygalacturonase gene of Ustilago maydis and characterization of the encoded enzyme. J Basic Microbiol. 54:340–349.
- Chang PK, Todd RB. 2004. Metabolic pathway regulation. In: Arora DK, editor. Handbook of fungal biotechnology. New York (NY): Marcel Dekker, Inc; p. 25–37.
- Cheung HYK, Donaldson ME, Spence KL, Fetsch JLO, Harrison MC, Saville BJ. Forthcoming 2017. Zfp1, a Zn(2)Cys(6) transcription factor, plays a key role in the virulence and pathogenesis of Ustilago maydis through the regulation of effector gene expression. Mol Plant Microbe Interact.
- Cho Y, Kim K-H, LaRota M, Scott D, Santopietro G, Callihan M, Mitchell TK, Lawrence CB. 2009. Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol Microbiol. 72:1316–1333.
- Chou H-M, Bundock N, Rolfe SA, Scholes JD. 2000. Infection of Arabidopsis thaliana leaves with Albugo candida (white blister rust) causes a reprogramming of host metabolism. Mol Plant Pathol. 1:99–113.
- Chow C-M, Yagüe E, Raguz S, Wood DA, Thurston CF. 1994. The cel3 gene of Agaricus bisporus codes for a modular cellulase and is transcriptionally regulated by the carbon source. Appl Environ Microbiol. 60:2779–2785.
- Couturier M, Navarro D, Olivé C, Chevret D, Haon M, Favel A, Lesage-Meessen L, Henrissat B, Coutinho PM, Berrin J-G. 2012. Post-genomic analyses of fungal lignocellulosic biomass degradation reveal the unexpected potential of the plant pathogen Ustilago maydis. BMC Genom. 13:57.
- Culleton H, McKie V, de Vries RP. 2013. Physiological and molecular aspects of degradation of plant polysaccharides by fungi: what have we learned from Aspergillus? Biotechnol J. 8:884–894.
- de Vries RP. 2003. Regulation of Aspergillus genes encoding plant cell wall polysaccharide-degrading enzymes; relevance for industrial production. Appl Microbiol Biotechnol. 61:10–20.
- de Vries RP, Visser J. 2001. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev. 65:497–522.
- DeVit MJ, Waddle JA, Johnston M. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 8:1603–1618.
- Ding S-J, Ge W, Buswell JA. 2001. Endoglucanase I from the edible straw mushroom, Volvariella volvacea: purification, characterization, cloning and expression. Eur J Biochem. 268:5687–5695.
- Divon HH, Fluhr R. 2007. Nutrition acquisition strategies during fungal infection of plants. FEMS Microbiol Lett. 266:65–74.
- Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I, et al. 2011. Metabolic priming by a secreted fungal effector. Nature. 478:395–398.
- Doehlemann G, Molitor F, Hahn M. 2005. Molecular and functional characterization of a fructose specific transporter from the gray mold fungus Botrytis cinerea. Fungal Genet Biol. 42:601–610.
- Doehlemann G, Wahl R, Horst RJ, Voll LM, Usadel B, Poree F, Stitt M, Pons-Kühnemann J, Sonnewald U, Kahmann R, et al. 2008b. Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis. Plant J. 56:181–195.
- Doehlemann G, Wahl R, Vranes M, de Vries RP, Kämper J, Kahmann R. 2008a. Establishment of compatibility in the Ustilago maydis/maize pathosystem. J Plant Physiol. 165:29–40.
- Doidy J, Grace E, Kühn C, Simon-Plas F, Casieri L, Wipf D. 2012. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 17:413–422.
- Dolezal AL, Shu X, OBrian GR, Nielsen DM, Woloshuk CP, Boston RS, Payne GA. 2014. Aspergillus flavus infection induces transcriptional and physical changes in developing maize kernels. Front Microbiol. 5:384.
- Dowzer CE, Kelly JM. 1991. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 11:5701–5709.
- Doyle CE, Cheung HYK, Spence KL, Saville BJ. 2016. Unh1, an Ustilago maydis Ndt80-like protein, controls completion of tumour maturation, teliospore development, and meiosis. Fungal Genet Biol. 94:54–68.
- Eckardt NA. 2017. The plant cell reviews plant immunity: receptor-like kinases, ROS-RLK crosstalk, quantitative resistance, and the growth/defense trade-off. Plant Cell. 29:601–602.
- Feng J, Zhang H, Strelkov SE, Hwang S-F. 2014. The LmSNF1 gene is required for pathogenicity in the canola blackleg pathogen Leptosphaeria maculans. PLoS One. 9:e92503.
- Fernandez J, Marroquin-Guzman M, Wilson RA. 2014. Mechanisms of nutrient acquisition and utilization during fungal infections of leaves. Annu Rev Phytopathol. 52:155–174.
- Fotopoulos V, Gilbert MJ, Pittman JK, Marvier AC, Buchanan AJ, Sauer N, Hall JL, Williams LE. 2003. The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, Atβfruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol. 132:821–829.
- Gancedo JM. 1998. Yeast carbon catabolite repression. Microbiol Mol Bio Rev. 62:334–361.
- Geiser E, Reindl M, Blank LM, Feldbrügge M, Wierckx N, Schipper K. 2016. Activating intrinsic CAZymes of the smut fungus Ustilago maydis for the degradation of plant cell wall components. Appl Environ Microbiol. 82:5174–5185.
- Geiser E, Wierckx N, Zimmermann M, Blank LM. 2013. Identification of an endo-1,4-beta-xylanase of Ustilago maydis. BMC Biotechnol. 13:59.
- Grabber JH, Ralph J, Hatfield RD. 2000. Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J Agric Food Chem. 48:6106–6113.
- Hayes MA, Feechan A, Dry IB. 2010. Involvement of abscisic acid in the coordinated regulation of a stress-inducible hexose transporter (VvHT5) and a cell wall invertase in grapevine in response to biotrophic fungal infection. Plant Physiol. 153:211–221.
- Heath MC. 1998. Apoptosis, programmed cell death and the hypersensitive response. Eur J Plant Pathol. 104:117–124.
- Heisterüber D, Schulte P, Moerschbacher BM. 1994. Soluble carbohydrates and invertase activity in stem rust-infected, resistant and susceptible near-isogenic wheat leaves. Physiol Mol Plant Pathol. 44:111–123.
- Helber N, Wippel K, Sauer N, Schaarschmidt S, Hause B, Requena N. 2011. A versatile monosaccharide transporter that operates in the arbuscular mycorrhizal fungus Glomus sp is crucial for the symbiotic relationship with plants. Plant Cell. 23:3812–3823.
- Hemetsberger C, Herrberger C, Zechmann B, Hillmer M, Doehlemann G. 2012. The Ustilago maydis effector Pep1 suppresses plant immunity by inhibition of host peroxidase activity. PLoS Pathog. 8:e1002684.
- Hemetsberger C, Mueller AN, Matei A, Herrberger C, Hensel G, Kumlehn J, Mishra B, Sharma R, Thines M, Hückelhoven R, et al. 2015. The fungal core effector Pep1 is conserved across smuts of dicots and monocots. New Phytol. 206:1116–1126.
- Herbers K, Meuwly P, Métraux J-P, Sonnewald U. 1996. Salicylic acid- independent induction of pathogenesis-related protein transcripts by sugars is dependent on leaf developmental stage. FEBS Lett. 397:239–244.
- Horst RJ, Doehlemann G, Wahl R, Hofmann J, Schmiedl A, Kahmann R, Kämper J, Voll LM. 2010. A model of Ustilago maydis leaf tumor metabolism. Plant Signal Behav. 5:1446–1449.
- Horst RJ, Engelsdorf T, Sonnewald U, Voll LM. 2008. Infection of maize leaves with Ustilago maydis prevents establishment of C4 photosynthesis. J Plant Physiol. 165:19–28.
- Jeffries TW. 1983. Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng Biotechnol. 27:1–32.
- Kämper J, Kahmann R, Bölker M, Ma L-J, Brefort T, Saville BJ, Banuett F, Kronstad JW, Gold SE, Müller O, et al. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature. 444:97–101.
- Kaupert Neto AA, Borin GP, Goldman GH, Damásio AR, Oliveira JV. 2016. Insights into the plant polysaccharide degradation potential of the xylanolytic yeast Pseudozyma brasiliensis. FEMS Yeast Res. 16:fov117.
- Kelly JM. 2004. The regulation of carbon metabolism in filamentous fungi. In: Brambl R, Marzluf GA, editors. The mycota III. Biochemistry and molecular biology. Berlin-Heidleberg: Springer-Verlag; p. 386–401.
- Kobakhidze A, Asatiani M, Kachlishvili E, Elisashvili V. 2016. Induction and catabolite repression of cellulase and xylanase synthesis in the selected white-rot basidiomycetes. Ann Agrar Sci. 14:169–176.
- Kolpak FJ, Blackwell J. 1976. Determination of the structure of cellulose II. Macromolecules. 9:273–278.
- Kretschmer M, Croll D, Kronstad JW. 2016. Maize susceptibility to Ustilago maydis is influenced by genetic and chemical perturbation of carbohydrate allocation. Mol Plant Pathol. doi:10.1111/mpp.12486
- Kruger WM, Pritsch C, Chao S, Muehlbauer GJ. 2002. Functional and comparative bioinformatic analysis of expressed genes from wheat spikes infected with Fusarium graminearum. Mol Plant Microbe Interact. 15:445–455.
- Kubicek CP, Starr TL, Glass NL. 2014. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol. 52:427–451.
- Kushalappa AC, Yogendra KN, Karre S. 2016. Plant innate immune response: qualitative and quantitative resistance. Crit Rev Plant Sci. 35:38–55.
- Lammens W, Le Roy K, Schroeven L, Van Laere A, Rabijns A, Van den Ende W. 2009. Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. J Exp Bot. 60:727–740.
- Lanver D, Berndt P, Tollot M, Naik V, Vranes M, Warmann T, Münch K, Rössel N, Kahmann R. 2014. Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathog. 10:e1004272.
- Lemoine R, La Camera S, Atanassova R, Dédaldéchamp F, Allario T, Pourtau N, Bonnemain J-L, Laloi M, Coutos-Thévenot P, Maurousset L, et al. 2013. Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci. 4:272.
- Levasseur A, Lomascolo A, Chabrol O, Ruiz-Dueñas FJ, Boukhris-Uzan E, Piumi F, Kües U, Ram AFJ, Murat C, Haon M, et al. 2014. The genome of the white-rot fungus Pycnoporus cinnabarinus: a basidiomycete model with a versatile arsenal for lignocellulosic biomass breakdown. BMC Genom. 15:486.
- Lingner U, Münch S, Deising HB, Sauer N. 2011. Hexose transporters of a hemibiotrophic plant pathogen: functional variations and regulatory differences at different stages of infection. J Biol Chem. 286:20913–20922.
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–495.
- Mäkelä MR, Donofrio N, de Vries RP. 2014. Plant biomass degradation by fungi. Fungal Genet Biol. 72:2–9.
- Malinovsky FG, Fangel JU, Willats WGT. 2016. The role of the cell wall in plant immunity. Front Plant Sci. 5:178.
- Martínez-Soto D, Robledo-Briones AM, Estrada-Luna AA, Ruiz-Herrera J. 2013. Transcriptomic analysis of Ustilago maydis infecting Arabidopsis reveals important aspects of the fungus pathogenic mechanisms. Plant Signal Behav. 8:e25059.
- Matei A, Doehlemann G. 2016. Cell biology of corn smut disease – Ustilago maydis as a model for biotrophic interactions. Curr Opin Microbiol. 34:60–66.
- Méndez-Morán L, Reynaga-Peña CG, Springer PS, Ruiz-Herrera J. 2005. Ustilago maydis infection of the nonnatural host Arabidopsis thaliana. Phytopathology. 95:480–488.
- Miwa T, Takagi Y, Shinozaki M, Yun C-W, Schell WA, Perfect JR, Kumagai H, Tamaki H. 2004. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot Cell. 3:919–931.
- Morkunas I, Ratajczak L. 2014. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol Plant. 36:1607–1619.
- Morrison EN, Emery RJN, Saville BJ. 2015. Phytohormone involvement in the Ustilago maydis–Zea mays pathosystem: relationships between abscisic acid and cytokinin levels and strain virulence in infected cob tissue. PLoS One. 10:e0130945.
- Morrison EN, Emery RJN, Saville BJ. 2017. Fungal derived cytokinins are necessary for normal Ustilago maydis infection of maize. Plant Pathol. 66:726–742.
- Mueller AN, Ziemann S, Treitschke Aßmann D, Doehlemann G. 2013. Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector pit2. Plos Pathog. 9:e1003177.
- Mueller O, Kahmann R, Aguilar G, Trejo-Aguilar B, Wu A, de Vries RP. 2008. The secretome of the maize pathogen Ustilago maydis. Fungal Genet Biol. 45:S63–70.
- Nadal M, Garcia-Pedrajas MD, Gold SE. 2010. The snf1 gene of Ustilago maydis acts as a dual regulator or cell wall degrading enzymes. Phytopathology. 100:1364–1372.
- Nahas E, Waldemarin MM. 2002. Control of amylase production and growth characteristics of Aspergillus ochraceus. Rev Latinoam Microbiol. 44:5–10.
- Nehlin JO, Carlberg M, Ronne H. 1991. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 10:3373–3377.
- Ohta A. 1997. Ability of ectomycorrhizal fungi to utilize starch and related substrates. Mycoscience. 38:403–408.
- Oliveira JV, Borges TA, Corrêa Dos Santos RA, Freitas LF, Rosa CA, Goldman GH, Riaño-Pachón DM. 2014. Pseudozyma brasiliensis sp. nov., a xylanolytic, ustilaginomycetous yeast species isolated from an insect pest of sugarcane roots. Int J Syst Evol Microbiol. 64:2159–2168.
- Onishi N, Tanaka T. 1996. Purification and properties of a galacto- and gluco-oligosaccharide-producing β-glycosidase from Rhodotorula minuta IFO879. J Ferment Bioeng. 82:439–443.
- Ospina-Giraldo MD, Mullins E, Kang S. 2003. Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis. Curr Genet. 44:49–57.
- Ostling J, Carlberg M, Ronne H. 1996. Functional domains in the Mig1 repressor. Mol Cell Biol. 16:753–761.
- Ostling J, Ronne H. 1998. Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem. 252:162–168.
- Papamichos-Chronakis M, Gligoris T, Tzamarias D. 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5:368–372.
- Parrent JL, James TY, Vasaitis R, Taylor AF. 2009. Friend or foe? Evolutionary history of glycoside hydrolase family 32 genes encoding for sucrolytic activity in fungi and its implications for plant-fungal symbioses. BMC Evol Biol. 9:148.
- Pons T, Olmea O, Chinea G, Beldarraín A, Márquez G, Acosta N, Rodríguez L, Valencia A. 1998. Structural model for family 32 of glycosyl-hydrolase enzymes. Proteins Struct Funct Genet. 33:383–395.
- Ries LN, Beattie SR, Espeso SA, Cramer RA, Goldman GH. 2016. Diverse regulation of the CreA carbon catabolite repressor in Aspergillus nidulans. Genetics. 203:335–352.
- Rolland F, Baena-Gonzalez E, Sheen J. 2006. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 57:675–709.
- Ronne H. 1995. Glucose repression in fungi. Trends Genet. 11:12–17.
- Ruijter GJ, Visser J. 1997. Carbon repression in Aspergilli. FEMS Microbiol Lett. 151:103–114.
- Rytioja J, Hildén K, Yuzon J, Hatakka A, de Vries RP, Mäkelä MR. 2014. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol Mol Bio Rev. 78:614–649.
- Sauer N, Ludwig A, Knoblauch A, Rothe P, Gahrtz M, Klebl F. 2004. AtSUC8 and AtSUC9 encode functional sucrose transporters, but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant J. 40:120–130.
- Saville BJ, Donaldson ME, Doyle CE. 2012. Investigating host induced meiosis in a fungal plant pathogen. In: Swan A, editor. Meiosis – molecular mechanisms and cytogenetic diversity. Rijeka: InTech; p. 411–460.
- Schauwecker F, Wanner G, Kahmann R. 1995. Filament-specific expression of a cellulase gene in the dimorphic fungus Ustilago maydis. Biol Chem Hoppe Seyler. 376:617–625.
- Schuler D, Wahl R, Wippel K, Vranes M, Münsterkötter M, Sauer N, Kämper J. 2015. Hxt1, a monosaccharide transporter and sensor required for virulence of the maize pathogen Ustilago maydis. New Phytol. 206:1086–1100.
- Schuster M, Schweizer G, Reissmann S, Kahmann R. 2016. Genome editing in Ustilago maydis using the CRISPR–Cas system. Fungal Genet Biol. 89:3–9.
- Seiboth B, Metz B. 2011. Fungal arabinan and L-arabinose metabolism. Appl Microbiol Biotechnol. 89:1665–1673.
- Shewale JG, Sadana J. 1981. Purification, characterization, and properties of β-glucosidase enzymes from Sclerotium rolfsii. Arch Biochem Biophys. 207:185–196.
- Siemens J, González M-C, Wolf S, Hofmann C, Greiner S, Du Y, Rausch T, Roitsch T, Ludwig-Müller J. 2011. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol Plant Pathol. 12:247–262.
- Skibbe DS, Doehlemann G, Fernandes J, Walbot V. 2010. Maize tumours caused by Ustilago maydis require organ-specific genes in host and pathogen. Science. 328:89–92.
- Smith DG, Garcia-Pedrajas MD, Gold SE, Perlin MH. 2003. Isolation and characterization from pathogenic fungi of genes encoding ammonium permeases and their roles in dimorphism. Mol Microbiol. 50:259–275.
- Strauss J, Horvath HK, Abdallah BM, Kindermann J, Mach RL, Kubicek CP. 1999. The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol Microbiol. 32:169–178.
- Sutton P, Gilbert M, Williams L, Hall JL. 2007. Powdery mildew infection of wheat leaves changes host solute transport and invertase activity. Physiol Plant. 129:787–795.
- Talbot NJ. 2010. Living the sweet life: how does a plant pathogenic fungus acquire sugar from plants? PLoS Biol. 8:e1000308.
- Tanaka S, Brefort T, Neidig N, Djamei A, Kahnt J, Vermerris W, Koenig S, Feussner K, Feussner I, Kahmann R. 2014. A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. eLife. 3:e01355.
- Tauzin AS, Giardina T. 2014. Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front Plant Sci. 5:293.
- Tetlow IJ, Farrar JF. 1992. Sucrose-metabolizing enzymes from leaves of barley infected with brown rust (Puccinia hordei Otth). New Phytol. 120:475–480.
- Todd RB, Zhou M, Ohm RA, Leeggangers HACF, Visser L, de Vries RP. 2014. Prevalence of transcription factors in ascomycete and basidiomycete fungi. BMC Genom. 15:214.
- Tollot M, Assmann D, Becker C, Altmüller J, Dutheil JY, Wegner C-E, Kahmann R. 2016. The WOPR protein Ros1 is a master regulator of sporogenesis and late effector gene expression in the maize pathogen Ustilago maydis. PLoS Pathog. 12:e1005697.
- Tonukari NJ, Scott-Craig JS, Walton JD. 2000. The Cochliobolus carbonum SNF1 gene is required for cell wall–degrading enzyme expression and virulence on maize. Plant Cell. 12:237–248.
- van den Brink J, de Vries RP. 2011. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 91:1477–1492.
- Van der Nest MA, Steenkamp ET, McTaggart AR, Trollip C, Godlonton T, Sauerman E, Roodt D, Naidoo K, Coetzee MPA, Wilken PM, et al. 2015. Saprophytic and pathogenic fungi in the Ceratocystidaceae differ in their ability to metabolize plant-derived sucrose. BMC Evol Biol. 15:273.
- Vanden Wymelenberg A, Minges P, Sabat G, Martinez D, Aerts A, Salamov A, Grigoriev I, Shapiro H, Putnam N, Belinky P, et al. 2006. Computational analysis of the Phanerochaete chrysosporium v2.0 genome database and mass spectrometry identification of peptides in ligninolytic cultures reveal complex mixtures of secreted proteins. Fungal Genet Biol. 43:343–356.
- Voegele RT, Struck C, Hahn M, Mendgen K. 2001. The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc Natl Acad Sci USA. 98:8133–8138.
- Voegele RT, Wirsel S, Möll U, Lechner M, Mendgen K. 2006. Cloning and characterization of a novel invertase from the obligate biotroph Uromyces fabae and analysis of expression patterns of host and pathogen invertases in the course of infection. Mol Plant Microbe Interact. 19:625–634.
- Vogel J. 2008. Unique aspects of the grass cell wall. Curr Opin Plant Biol. 11:301–307.
- Voll LM, Horst RJ, Voitsik AM, Zajic D, Samans B, Pons-Kühnemann J, Doehlemann G, Münch S, Wahl R, Molitor A, et al. 2011. Common motifs in the response of cereal primary metabolism to fungal pathogens are not based on similar transcriptional reprogramming. Front Plant Sci. 2:39.
- Wahl R, Wippel K, Goos S, Kämper J, Sauer N. 2010. A novel high-affinity sucrose transporter is required for virulence of the plant pathogen Ustilago maydis. PLoS Biol. 8:e1000303.
- Wilson WA, Hawley SA, Hardie DG. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 6:1426–1434.
- Wippel K, Wittek A, Hedrich R, Sauer N. 2010. Inverse pH regulation of plant and fungal sucrose transporters: a mechanism to regulate competition for sucrose at the host/pathogen interface? PLoS One. 5:e12429.
- Yi M, Park J-H, Ahn J-H, Lee Y-H. 2008. MoSNF1 regulates sporulation and pathogenicity in the rice blast fungus Magnaporthe oryzae. Fungal Genet Biol. 45:1172–1181.
- Zeilinger S, Gupta VK, Dahms TES, Silva RN, Singh HB, Upadhyay RS, Gomes EV, Tsui CK-M, Nayak SC. 2016. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol Rev. 40:182–207.
- Zhang Z, Henderson C, Perfect E, Carver TL, Skamnioti P, Gurr SJ. 2005. Of genes and genomes, needles and haystacks: Blumeria graminis and functionality. Mol Plant Pathol. 6:561–575.
- Zhao Z, Liu H, Wang C, Xu J-R. 2013. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 14:274.
