Abstract
Due to health risks and economic losses associated with mycotoxins produced by Fusarium species, there is a compelling need for an improved understanding of these fungi from across diverse perspectives and disciplinary approaches. In this article, we provide a transdisciplinary overview of: (i) Fusarium phylogenetics; (ii) linkages between mycotoxin biosynthetic gene clusters and chemical structures; (iii) biotransformation of mycotoxins to reduce toxicity; (iv) Fusarium population biology; (v) genomics of secondary metabolite production; and (vi) mycotoxigenic fusaria in a phytobiomes context. Phylogenetic studies have made tremendous progress in delineating the species that comprise the genus Fusarium, many of which are morphologically cryptic. Accurate species identification and a thorough understanding of the distribution of mycotoxin biosynthetic genes among those species will facilitate control of mycotoxin contamination. The biochemical pathways leading to the formation of several Fusarium mycotoxins have been elegantly linked with the genes responsible for each chemical transformation during synthesis, and for most structural differences among chemotypes. Screens for the biotransformation of mycotoxins have led to the description of chemical modifications that impact bioactivity and have implications for monitoring and testing of the food supply. Population biology studies have revealed the potential for introductions of foreign genotypes to alter regional populations of mycotoxigenic fusaria. Genomic analyses have begun to reveal the complex evolutionary history of the genes responsible for mycotoxin production, both across and within lineages. Improved understanding of how climate variability impacts plant–Fusarium interactions and mycotoxin accumulation is necessary for effective plant resistance. Additionally, improved understanding of interactions between Fusarium and other members of crop microbiomes is expected to produce novel strategies for limiting disease and mycotoxin accumulation.
Résumé
À cause des risques pour la santé et des pertes économiques associés aux mycotoxines produites par des espèces du genre Fusarium, le besoin de mieux comprendre ces champignons est devenu impérieux, et ce, sous divers angles et en fonction de diverses approches multidisciplinaires. Dans cet article, nous offrons un aperçu transdisciplinaire: (i) de la phylogénétique de Fusarium; (ii) des relations entre les groupes de gènes biosynthétiques des mycotoxines et les structures chimiques; (iii) de la biotransformation des mycotoxines afin d’en réduire la toxicité; (iv) de la biologie des populations de Fusarium; (v) de la génomique de la production des métabolites secondaires; et (vi) des espèces mycotoxigéniques du genre Fusarium dans le contexte du phytobiome. Les études phylogénétiques ont fait d’immenses progrès en déterminant les espèces qui constituent le genre Fusarium, dont plusieurs sont morphologiquement cryptiques. L’identification précise des espèces et une compréhension approfondie de la distribution des gènes biosynthétiques des mycotoxines chez ces dernières faciliteront la lutte contre la contamination par les mycotoxines. Les voies biochimiques menant à la formation de plusieurs mycotoxines de Fusarium ont été reliées de manière sophistiquée aux gènes responsables de chaque transformation chimique durant la synthèse et de la plupart des différences structurales chez les chimiotypes. Le criblage concernant la biotransformation des mycotoxines a permis de décrire les modifications chimiques qui influencent la bioactivité et qui ont des incidences sur le suivi et l’analyse de l’approvisionnement alimentaire. Les études sur la biologie des populations ont mis en évidence les possibilités qu’ont les génotypes étrangers introduits d’altérer les populations régionales d’espèces mycotoxigéniques du genre Fusarium. Les analyses génomiques ont commencé à révéler l’histoire évolutionnaire complexe des gènes responsables de la production des mycotoxines, et ce, non seulement entre les lignées, mais au sein de celles-ci également. Il importe de comprendre en profondeur l’influence de la variabilité climatique sur les interactions plante–Fusarium et l’accumulation des mycotoxines afin que les plantes puissent afficher une résistance efficace. En outre, une compréhension approfondie des interactions entre Fusarium et les autres plantes cultivées du microbiome devrait contribuer à élaborer de nouvelles stratégies visant à faire obstacle à la maladie et à prévenir l’accumulation de mycotoxines.
Introduction
The genus Fusarium (Ascomycota; Sordariomycetes; Hypocreales) is ubiquitous in agronomic systems around the world and includes many economically important plant pathogens. Plant diseases caused by Fusarium species include seedling blights and root rots (Bakker et al., Citation2016), vascular wilts (Michielse & Rep, Citation2009), diseases of reproductive tissues and developing seeds (Kazan et al., Citation2012), and storage diseases (Gachango et al., Citation2012).
Collectively, Fusarium species (fusaria) possess the genetic ability to produce hundreds of structurally diverse secondary metabolites, most of which have poorly understood or entirely unknown ecological functions (Ma et al., Citation2013; Hansen et al., Citation2015; Brown & Proctor, Citation2016; Niehaus et al., Citation2016; Kim et al., Citation2017). These metabolites include several toxins that act as virulence factors related to plant disease development (Proctor et al., Citation1995). Of even greater concern, however, are the health impacts for humans and livestock that consume grains contaminated with mycotoxins (Pestka, Citation2010; Wu et al., Citation2014).
Among the Fusarium mycotoxins of primary concern are the trichothecenes, fumonisins and zearalenone. Trichothecenes are produced by Fusarium graminearum Schwabe and its close relatives, and are associated with fusarium head blight of small grains as well as ear rot of corn (Zea mays L.). Trichothecenes are epoxide-containing sesquiterpenoid compounds that play a significant role in pathogen virulence in planta, likely due to their ability to inhibit eukaryotic protein synthesis (Cundliffe et al., Citation1974; Bai et al., Citation2001). Zearalenone is an oestrogen mimic (Kowalska et al., Citation2016) that is produced by F. graminearum and related species within the Fusarium sambucinum species complex. Zearalenone is not required for disease development on wheat (Triticum aestivum L.) (Munkvold, Citation2017). Fumonisins are produced by Fusarium verticillioides (Sacc.) Nirenberg and some of its close relatives, and are associated with ear rot of corn. Fumonisins are polyketide derived mycotoxins that are not required for disease of maize (Desjardins & Plattner, Citation2000), but have health impacts related to kidney and liver toxicity and neural tube defects and are probable carcinogens due likely to their ability to disrupt sphingolipid biosynthesis (Stockmann-Juvala & Savolainen, Citation2008).
Losses to the agricultural economy that are attributable to mycotoxigenic Fusarium spp. are valued at hundreds of millions of dollars per year at regional scales (Windels, Citation2000; Nganje et al., Citation2004) and likely reach costs of billions of dollars per year globally. Due to the health and economic costs of mycotoxins produced by Fusarium spp., there is a compelling need to study these fungi from across diverse perspectives and disciplinary approaches.
In this mini-review, we provide a transdisciplinary overview of: (i) Fusarium phylogenetics; (ii) linkages between mycotoxin biosynthetic gene clusters and chemical structures; (iii) biotransformation of mycotoxins to reduce toxicity; (iv) Fusarium population biology; (v) genomics of secondary metabolite production; and (vi) mycotoxigenic fusaria in a phytobiomes context.
Phylogenetics and classification of mycotoxigenic Fusarium
First erected by Link (Citation1809), and subsequently validated by Fries (Citation1821), the genus Fusarium has undergone many taxonomic revisions over the past two centuries. Following the movement to a ‘one fungus, one name’ system of nomenclature, the research community expressed strong support for circumscribing the limits of Fusarium to preserve historical use of the name (Geiser et al., Citation2013). This circumscription was supported by molecular phylogenetic analyses of partial nucleotide sequences for RNA polymerase genes RPB1 and RPB2, which provided a well-supported phylogenetic hypothesis of evolutionary relationships within the genus (O’Donnell et al., Citation2013). Results of this analysis resolved 20 monophyletic species complexes and nine monotypic lineages. Two of these monotypic lineages, based on newly discovered species, are now recognized as species complexes (Laurence et al., Citation2011; Zhou et al., Citation2016).
Clearly delineating species within the genus Fusarium, and defining the distribution of mycotoxigenic phenotypes among those species, will greatly facilitate progress toward effective control of plant diseases and mycotoxin contamination of food and feed. To support this effort, more than 14 000 phylogenetically diverse Fusarium strains have been accessioned in the Agricultural Research Service (ARS) Culture Collection (Peoria, IL), where they are available for distribution upon request. Many of these strains have been genetically characterized using Genealogical Concordance Phylogenetic Species Recognition (GCPSR; see Sarver et al., Citation2011), a robust method for identifying species boundaries (Taylor et al., Citation2000). To date, GCPSR-based studies indicate that approximately half of the ~300 phylogenetically distinct species-level Fusarium lineages represented within the ARS Culture Collection are not distinguishable from other species by morphological traits, and are currently unnamed (Aoki et al., Citation2014).
In an effort to enable and promote accurate, species-level identification of Fusarium isolates, two web-accessible DNA sequence databases (i.e. FUSARIUM-ID and Fusarium MLST) were developed (Geiser et al., Citation2004; Crous et al., Citation2015; O’Donnell et al., Citation2015). These databases are frequently updated with sequence data from newly characterized fusaria. To date, portions of three protein coding genes (translation elongation factor, TEF1; RPB1; RPB2) have been shown to be phylogenetically informative at or near the species level across the breadth of Fusarium (Geiser et al., Citation2004; O’Donnell et al., Citation2013). In contrast to GenBank, where many sequences assigned to Fusarium are misidentified (O’Donnell et al., Citation2015), all of the sequences in the two Fusarium-specific databases were derived from reference strains that can be obtained for additional study via the ARS Culture Collection (https://nrrl.ncaur.usda.gov/), the Fusarium Research Center at Pennsylvania State University (http://www.fusariumdb.org/) or the Westerdijk Fungal Biodiversity Institute (http://www.westerdijkinstitute.nl/; formerly CBS-KNAW).
Genetics and biosynthesis of Fusarium mycotoxins
Sustained research investments have provided tremendous insights into the genetics and biochemistry underlying the biosynthesis of most Fusarium mycotoxins, as well as the genetic bases for chemotype variation among strains. Here, we use trichothecene biosynthesis to illustrate the state of knowledge (), although similar levels of understanding exist for other mycotoxins (e.g. Kim et al., Citation2005; Alexander et al., Citation2009; Uhlig et al., Citation2012; Niehaus et al., Citation2013). From the primary metabolite farnesyl diphosphate, 7-10 enzymatic modifications lead to synthesis of trichothecene mycotoxins. Each enzymatic step in this process has been linked to a particular trichothecene biosynthetic (TRI) gene within or outside of the primary TRI cluster (). The cluster includes additional genes that encode regulatory proteins (TRI6, TRI10), a transporter (TRI12), and proteins of unknown functions (TRI9, TRI14) (Kimura et al., Citation2007; Alexander et al., Citation2009).
Fig. 1. (Colour online) Illustration of the biosynthetic pathway leading to production of trichothecenes, with linkages shown to the corresponding gene cluster for three generically represented Fusarium species. (Chemotype, and corresponding genotype, differences can be found within species.) Labels at the arrows indicate the TRI gene whose product enacts the indicated biotransformation. DON = deoxynivalenol; 3-ADON = 3-acetyl-deoxynivalenol; 15-ADON = 15-acetyl-deoxynivalenol; NIV = nivalenol; NX-2 = NX-2 toxin; NX-3 = NX-3 toxin; T-2 = T-2 toxin. Coloured polygons indicate the presence and orientation of TRI genes within a genome. Gene lengths are not drawn to scale. Ψ indicates a pseudogenized gene. Gene symbols are colour-coded by functional category, as indicated in the legend. The inset figure depicts a generic trichothecene backbone structure, and indicates the conventional numbering of atom positions.
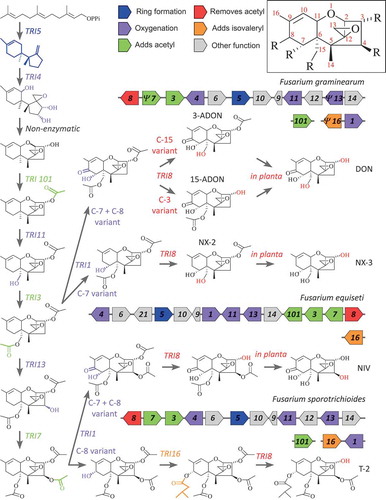
In some cases, chemotype variation results from differences in the presence and absence of biosynthetic genes. For instance, TRI16 is responsible for addition of a five-carbon moiety (isovalerate) to the oxygen at C-8 of the trichothecene molecule during biosynthesis of T-2 toxin (; Brown et al., Citation2003; Peplow et al., Citation2003). TRI16 is present and functional in species that produce T-2 toxin (e.g. Fusarium sporotrichioides Sherb.), but is absent or pseudogenized in species (e.g. F. graminearum) that produce trichothecene variants like nivalenol (NIV) and deoxynivalenol (DON), which lack an isovalerate moiety (; McCormick et al., Citation2004; Vanheule et al., Citation2016). Similarly, the presence or absence of a functional TRI13 gene is responsible for the DON and NIV chemotype polymorphism observed within F. graminearum and related species (; Brown et al., Citation2002; Lee et al., Citation2002; Kimura et al., Citation2003), which has been maintained by a form of balancing selection acting directly on these chemotype differences (Ward et al., Citation2002).
In other cases, trichothecene chemotype variation results from differences in function of allelic variants of the same TRI gene. For instance, in most F. graminearum strains, TRI1 is responsible for trichothecene oxygenation at both C-7 and C-8, leading to formation of variants like DON or NIV (; McCormick et al., Citation2004). However, in some F. graminearum strains, TRI1 adds an hydroxyl group at C-7 only, which leads to formation of NX-2 and related toxins (; Varga et al., Citation2015). In contrast, the F. sporotrichioides TRI1 adds an hydroxyl group at C-8 only, leading to formation of T-2 toxin (; Brown et al., Citation2003; Meek et al., Citation2003). Similarly, different TRI8 alleles are responsible for 3-acetyl-deoxynivalenol (3-ADON) vs. 15-acetyl-deoxynivalenol (15-ADON) chemotypes in F. graminearum, depending on whether TRI8 de-acetylates C-15 or C-3, respectively (; Alexander et al., Citation2011).
As a result of this understanding that links genetic determinants with chemical modifications to mycotoxin structure, the particular mycotoxin variant (i.e. chemotype) produced by an unknown isolate or a novel species of Fusarium can be readily inferred using DNA-based methods. For instance, a multilocus genotyping assay was developed to simultaneously identify fusarium head blight pathogens and predict their trichothecene chemotypes (Ward et al., Citation2008). Similarly, mycotoxigenic potential can be predicted by mining genome sequences for mycotoxin biosynthesis genes (Edwards et al., Citation2016; Gräfenhan et al., Citation2016). Chemotype predictions inferred from DNA-based analyses can be confirmed via chemical analysis of culture broths or extraction from solid culture substrates (Aoki et al., Citation2015; Edwards et al., Citation2016; Gräfenhan et al., Citation2016), although actual production is not always observed in strains that have the necessary biosynthetic genes.
Thus, prior investments to generate detailed understandings of the relationship between genotype and chemotype facilitate rapid characterization of novel isolates and their toxigenic potential. It is also anticipated that this detailed mechanistic understanding of mycotoxin biosynthesis will suggest targets for more effective prevention or control of mycotoxin contamination.
Biotransformation of Fusarium mycotoxins
Biochemical modifications can substantially modulate the bioactivity of Fusarium mycotoxins, and both plants and microbes belonging to disparate taxa have been described as having the capacity for such enzymatic modifications (e.g. McCormick et al., Citation2012; He et al., Citation2015; Li et al., Citation2015). With sufficient understanding of these processes, there is the potential for practical, applied usage in mitigating economic losses due to mycotoxins. For instance, enzymatic cleaning of contaminated grain may reduce mycotoxin content sufficiently to allow for some uses of that grain. Alternately, enzymes that are able to reduce the toxicity of Fusarium mycotoxins may represent novel strategies for enhancing plant defences toward Fusarium pathogens, or improving the efficacy of biological control agents.
Several excellent reviews of microbial biotransformation of mycotoxins are available (Palumbo et al., Citation2008; Awad et al., Citation2010; McCormick, Citation2013; Vanhoutte et al., Citation2016). Screens for mycotoxin-biotransforming microorganisms have led to the discovery of several different chemical transformations of DON and T-2 toxin. These include de-epoxidation (Fuchs et al., Citation2002), epimerization (Ikunaga et al., Citation2011), deacylation (Young et al., Citation2007) and glucosylation (McCormick et al., Citation2012).
Trichothecene toxicity depends heavily upon the epoxide moiety of the molecule, and opening the epoxide ring dramatically reduces toxicity (Zhou et al., Citation2008). Because oxygen status is linked to redox potential, and epoxide ring opening is a reductive process, it is not surprising that most reports of this biotransformation have involved anaerobic organisms and culture conditions. Microbial strains or consortia displaying anaerobic DON de-epoxidation have originated from environments such as rumen fluid (Fuchs et al., Citation2002) and the digesta of fish (Guan et al., Citation2009) and poultry (Yu et al., Citation2010; Li et al., Citation2011).
The oxygen atom at the C-3 position is another portion of the trichothecene molecule that imparts substantial toxicity. Acetylation of the C-3 oxygen helps protect Fusarium from the toxic effects of the trichothecenes during biosynthesis (Kimura et al., Citation1998; McCormick et al., Citation1999), but toxins are deacetylated in infected plant tissues, increasing their bioactivity. Addition of glucose to the C-3 oxygen by plant glycosyltransferase enzymes can convert trichothecenes into less toxic glycosides (Lemmens et al., Citation2005; Lin et al., Citation2008; Wetterhorn et al., Citation2016). For instance, expression of a glucosyltransferase from barley (Hordeum vulgare L.) in wheat enhances the resistance of the wheat to the secondary spread of F. graminearum through the spike (Li et al., Citation2015). Microbial biotransformations at the C-3 position (formation of 3-epimer, 3-keto) have been reported from Gram-negative (Devosia Nakagawa, Sakane & Yokota) and Gram-positive (Nocardioides Prauser) bacteria (Ikunaga et al., Citation2011; Sato et al., Citation2012; He et al., Citation2015).
Yeasts in the Blastobotrys Klopotek (Trichomonascus Jackson) clade can also reduce toxicity of trichothecenes by C-3 acetylation, C-3 glucosylation, and removing side groups from the trichothecene molecule (McCormick et al., Citation2012). From a food safety risk perspective, such biotransformations of mycotoxins pose challenges to detection if the structural change alters the sensitivity of detection. For instance, an antibody test for the detection of T-2 toxin did not effectively detect the glucosylated form of the toxin (McCormick et al., Citation2015). This necessitated development of an antibody for glucosylated T-2 toxin (Maragos et al., Citation2013).
Additional screening for biotransformation may eventually lead to identification of enzymes that detoxify mycotoxins, which would be a boon for both plant protection and food safety. However, it will be a challenge to design screening approaches that avoid re-discovering already known biotransformations. It is not yet known whether the enzymatic machinery exists for the mineralization of trichothecenes and other Fusarium mycotoxins. It is expected that such metabolic capacity exists within soil microbial communities, as mycotoxins do not appear to accumulate in the environment. However, the description of the complete metabolism of trichothecenes awaits further research.
Population biology of mycotoxigenic fusaria
Analyses employing a combination of multilocus genotyping and neutral molecular markers enable large-scale analyses of the diversity, mycotoxigenic potential and population structure among fusaria (Gale et al., Citation2007; Ward et al., Citation2008; Gale et al., Citation2011; Bec et al., Citation2014; Liang et al., Citation2014; Kelly et al., Citation2015; Liang et al., Citation2015). In North America, these studies have revealed two dominant populations of F. graminearum, known as NA1 and NA2. These populations have distinct demographic histories; the NA1 population is genetically diverse and comprised of native isolates that typically possess the 15-ADON chemotype, whereas NA2 represents an invasive population that has undergone a bottleneck and is associated with the 3-ADON chemotype. The NA2 population has rapidly spread across major wheat-growing regions, and in recent decades has become dominant in the Upper Midwestern USA and in western and Maritime Canada (Liang et al., Citation2014; Kelly et al., Citation2015). However, such shifts have not occurred in eastern Canada and other parts of the USA, where the NA1 population remains dominant (Schmale et al., Citation2011; Bec et al., Citation2014; Kelly et al., Citation2015).
Very recently, F. graminearum isolates possessing the novel NX-2 chemotype (see for chemical structure), were found to be sympatric with NA1 and NA2 in southern Canada and the northern US (Liang et al., Citation2015; Varga et al., Citation2015; Kelly et al., Citation2016). NX-2-producing F. graminearum have undergone toxin diversification in response to changes in selection pressure acting on the cytochrome P450 enzyme encoded by TRI1. This example demonstrates that adaptive constraints on the molecular evolution of trichothecene genes may be population or niche-specific, and further indicates that mycotoxin chemotype differences may be important in niche adaptation (Kelly et al., Citation2016).
While the ecological factors underlying F. graminearum population dynamics remain largely unknown, it would seem that a complex adaptive landscape of regional selection pressures has influenced the distribution of fusarium head blight pathogen populations and mycotoxin chemotypes in North America. Such analyses of the population biology of mycotoxigenic Fusarium spp. can inform assessment of the risks posed by introductions of foreign pathogen genotypes, and can reveal geographic population structure, with implications for mycotoxin testing, fungicide resistance and plant germplasm evaluation.
Genomic analyses of secondary metabolite production
A rapidly increasing number of fully sequenced genomes has enabled comparative and functional genomics analyses aimed specifically at mycotoxins. Comparative genomic analysis of secondary metabolite biosynthetic gene clusters in diverse genomes can identify chemotype-specific gene clusters (Semeiks et al., Citation2014) and novel metabolites (Wiemann et al., Citation2013), as well as providing insights into the evolution and origins of the genes responsible for secondary metabolite biosynthesis (Sieber et al., Citation2014).
There is wide variation in the presence and absence of mycotoxin biosynthetic genes among Fusarium spp. (Brown & Proctor, Citation2016). Increasingly, there is also evidence for intra-species variation in gene content for mycotoxins or other secondary metabolites. For instance, the biosynthetic gene cluster for fusarin production is intact in some strains of Fusarium proliferatum (Matsush.) Nirenberg, while portions of the cluster, including critical genes for fusarin production, have been lost in other strains (Niehaus et al., Citation2016). Other taxa that have largely been considered non-mycotoxigenic have recently been shown to possess genes enabling the potential production of diverse secondary metabolites; for instance, genomic analyses of Fusarium avenaceum (Fr.) Sacc. revealed 26 polyketide synthase genes and 24 non-ribosomal peptide synthase genes, which likely confer the ability to produce the mycotoxins beauvericin/enniatins, fusarins and the fumonisin-like metabolite 2-amino-14,16-dimethyloctadecan-3-ol (Uhlig et al., Citation2005; Lysøe et al., Citation2014; Hansen et al., Citation2015; Brown & Proctor, Citation2016).
Diverse mechanisms have contributed to the distribution of mycotoxin biosynthetic genes, including vertical inheritance, gene loss, horizontal transfer and gene duplication (Proctor et al., Citation2009; Proctor et al., Citation2013; Niehaus et al., Citation2016). For instance, comparative genomic analyses have revealed a complex history of horizontal gene transfer and subsequent uneven degradation of the gene cluster responsible for the production of depudecin (Reynolds et al., Citation2017), a histone deacetylase inhibitor produced by diverse Ascomycota. While some fusaria retain an intact and functional depudecin gene cluster, in others one or more depudecin genes are non-functional (pseudogenized) or have been lost (deleted). Interestingly, two genes from the depudecin cluster tend to be preferentially retained; an efflux pump (DEP3), and a transcription factor gene (DEP6) that regulates DEP gene expression. This finding suggests that retention of DEP3 and DEP6 provides a selective advantage, perhaps by conferring resistance to exogenous depudecin or structurally similar metabolites (Reynolds et al., Citation2017).
Improved understanding of how biosynthetic gene cluster expression is regulated also has the potential to improve management of mycotoxigenic fusaria, and can be informed by genomic analyses. Most mycotoxin biosynthetic gene clusters include one or more genes coding for pathway-specific transcription factors, such as TRI6 and TRI10 in the trichothecene biosynthetic cluster (), or FUB10 and FUB12 in the fusaric acid biosynthetic cluster (Brown et al., Citation2015). However, other clusters, such as the fusarin gene cluster, do not include a pathway-specific regulatory gene (Niehaus et al., Citation2013). Expression of genes in the latter clusters is likely controlled directly by global regulators encoded by genes located outside the cluster (Brakhage, Citation2013). In contrast, global regulatory elements likely exercise less direct control over clusters that encode a pathway-specific transcription factor(s) (Studt et al., Citation2012; Niehaus et al., Citation2014), but more likely have indirect control via the transcription factor(s). These regulatory dynamics relate to the role of secondary metabolite production in the broader life history strategy of fusaria that produce them. For instance, global regulatory elements may be required for broad reshaping of gene expression during transition from one habitat to another, such as from saprotrophic growth to colonization of a living plant (Brown et al., Citation2014).
Finally, many secondary metabolite clusters of unknown function remain to be explored. We anticipate that these metabolites play a role not only in plant-microbe interactions, but also in species interactions with other microbes.
Mycotoxigenic fusaria in a phytobiomes context
The success of Fusarium pathogens, and the accumulation of associated mycotoxins, depends heavily on interactions with other organisms, and on environmental conditions. The concept of the phytobiome has been recently championed as an integrating idea that ties together plants, their environment, and the complex communities of organisms that interact with plants (Young & Kinkel, Citation2017). There remains much to understand regarding mycotoxin production by fusaria from a phytobiome perspective. For instance, since fumonisins do not appear to be required for virulence in planta (Desjardins & Plattner, Citation2000), do they instead play a more active role in competitive interactions with other microbes?
Previous crop rotations, tilling practices, and other agronomic practices, which also influence soil microbial communities, have been shown to influence crop diseases and mycotoxin contamination caused by Fusarium. Additionally, weather is a key factor driving the life-cycles of Fusarium and of other microorganisms within the phytobiome, and ultimately the host–pathogen interactions. Thus, to understand this complex ecological system, there must be simultaneous consideration of the host plant, the pathogenic Fusarium, and the broader microbiome, all in relation to the abiotic environmental context.
Changes in climate directly influence host defence responses and therefore host–pathogen interactions. Several recent studies investigating the influence of climatic variables on host plant defences against Fusarium infection and mycotoxin contamination have been published (Vaughan et al., Citation2014; Vaughan et al., Citation2016a; Vaughan et al., Citation2016b). Corn grown at an elevated atmospheric carbon dioxide (CO2) concentration was more susceptible to F. verticillioides proliferation, although it did not affect fumonisin contamination (Vaughan et al., Citation2014). Changes to atmospheric CO2 concentration had cascading effects on plant gene expression that led to a compromised defence response on one hand, but a reduction in host plant factors that induce mycotoxin production by F. verticillioides at the same time (Vaughan et al., Citation2014). However, the combination of both elevated CO2 concentration and drought stress resulted in even greater susceptibility of the plant to F. verticillioides proliferation and increased fumonisin contamination (Vaughan et al., Citation2016b).
While these results provide valuable information on direct host–pathogen interactions in the context of different climatic variables, many other indirect effects of elevated CO2 concentration and drought likely influence disease development and mycotoxin accumulation. For example, herbivore feeding damage is known to affect the susceptibility of host plants to Fusarium infection and mycotoxin contamination (Bowers et al., Citation2014). Weather variability also results in changes in herbivore populations and feeding behaviours. Recent evidence suggests that elevated CO2 concentration also reduces the production and release of volatile organic chemical signals that function in crop tri-trophic interactions (Block et al., Citation2017).
The potential impact of changing climate on the F. graminearum–wheat pathosystem was recently reviewed (Vaughan et al., Citation2016a). Abiotic pressures influence the major processes of the saprotrophic and pathogenic phases of the fusarium head blight disease cycle, including inoculum production. Shifts in host plant phenology due to changes in dominant annual weather patterns will also likely improve synchrony between pathogen inoculum production and host plant flowering, resulting in increases in disease development. Nevertheless, there remains considerable uncertainty around the potential for altered environmental conditions to influence mycotoxin contamination, particularly since so many unknowns still exist regarding the direct and indirect impact of climate changes on the many ecological dynamics that can influence host–pathogen interactions.
Recent research has used culture-independent techniques to semi-comprehensively describe the complement of microorganisms associated with plants that host mycotoxigenic fusaria (Karlsson et al., Citation2014; Nicolaisen et al., Citation2014; Grudzinska-Sterno et al., Citation2016; Hertz et al., Citation2016). These efforts have been complemented by culture-based efforts to collect isolates representative of the host plant microbiome (Yoshida et al., Citation2012; Comby et al., Citation2016). These efforts are expected to advance the goal of effective biological control of mycotoxigenic fusaria (Comby et al., Citation2017), whether through inoculation with antagonistic microbes or by means of managing the phytobiome to enhance pathogen suppression by the indigenous microbiome.
Conclusions
Mycotoxigenic fusaria pose serious barriers to human well-being, animal health and agricultural productivity. However, transdisciplinary study of these fungi and their secondary metabolites has produced powerful insights that will enable more effective control practices. Despite our growing understanding, it is clear that further research will continue to more accurately define the food safety risks associated with mycotoxins produced by Fusarium spp., and to illuminate the ecological factors that contribute to the success of these versatile and interesting organisms.
Disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer. This article was the work of US Government employees engaged in their official duties and is exempt from copyright.
Additional information
Funding
References
- Alexander NJ, McCormick SP, Waalwijk C, van der Lee T, Proctor RH. 2011. The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in Fusarium. Fungal Genet Biol. 48:485–495.
- Alexander NJ, Proctor RH, McCormick SP. 2009. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 28:198–215.
- Aoki T, O’Donnell K, Geiser DM. 2014. Systematics of key phytopathogenic Fusarium species: current status and future challenges. J Gen Plant Pathol. 80:189–201.
- Aoki T, Vaughan MM, McCormick SP, Busman M, Ward TJ, Kelly A, O’Donnell K, Johnston PR, Geiser DM. 2015. Fusarium dactylidis sp. nov., a novel nivalenol toxin-producing species sister to F. pseudograminearum isolated from orchard grass (Dactylis glomerata) in Oregon and New Zealand. Mycologia 107:409–418.
- Awad WA, Ghareeb K, Bohm J, Zentek J. 2010. Decontamination and detoxification strategies for the Fusarium mycotoxin deoxynivalenol in animal feed and the effectiveness of microbial biodegradation. Food Addit Contam. 27:510–520.
- Bai GH, Desjardins AE, Plattner RD. 2001. Deoxynivalenol-nonproducing Fusarium graminearum causes initial infection, but does not cause disease spread in wheat spikes. Mycopathologia 153:91–98.
- Bakker MG, Acharya J, Moorman TB, Robertson AE, Kaspar TC. 2016. The potential for cereal rye cover crops to host corn seedling pathogens. Phytopathology 106:591–601.
- Bec S, Ward T, Farman M, O’Donnell K, Hershman D, Van Sanford D, Vaillancourt LJ. 2014. Characterization of Fusarium strains recovered from wheat with symptoms of head blight in Kentucky. Plant Dis. 99:1622–1632.
- Block A, Vaughan MM, Christensen SA, Alborn HT, Tumlinson JH. 2017. Elevated carbon dioxide reduces emission of herbivore induced volatiles in Zea mays. Plant Cell Environ. 40:1725–1734.
- Bowers E, Hellmich R, Munkvold G. 2014. Comparison of fumonisin contamination using HPLC and ELISA methods in Bt and near-isogenic maize hybrids infested with European corn borer or Western bean cutworm. J Agric Food Chem. 62:6463–6472.
- Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 11:21–32.
- Brown DW, Busman M, Proctor RH. 2014. Fusarium verticillioides SGE1 is required for full virulence and regulates expression of protein effector and secondary metabolite biosynthetic genes. Mol Plant-Microbe Interact. 27:809–823.
- Brown DW, Lee S-H, Kim L-H, Ryu J-G, Lee S, Seo Y, Kim YH, Busman M, Yun S-H, Proctor RH, Lee T. 2015. Identification of a 12-gene fusaric acid biosynthetic gene cluster in Fusarium species through comparative and functional genomics. Mol Plant-Microbe Interact. 28:319–332.
- Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. 2002. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet Biol. 36:224–233.
- Brown DW, Proctor RH. 2016. Insights into natural products biosynthesis from analysis of 490 polyketide synthases from Fusarium. Fungal Genet Biol. 89:37–51.
- Brown DW, Proctor RH, Dyer RB, Plattner RD. 2003. Characterization of a Fusarium 2-gene cluster involved in trichothecene C-8 modification. J Agric Food Chem. 51:7936–7944.
- Comby M, Gacoin M, Robineau M, Rabenoelina F, Ptas S, Dupont J, Profizi C, Baillieul F. 2017. Screening of wheat endophytes as biological control agents against Fusarium head blight using two different in vitro tests. Microbiol Res. 202:11–20.
- Comby M, Lacoste S, Baillieul F, Profizi C, Dupont J. 2016. Spatial and temporal variation of cultivable communities of co-occurring endophytes and pathogens in wheat. Frontier Microbiol. 7:403.
- Crous PW, Robert VARG, Lombard L, Alejandra G, van Diepeningen A, O’Donnell K, Ward TJ. 2015. Fusarium MLST database. Available from http://www.westerdijkinstitute.nl/fusarium/.
- Cundliffe E, Cannon M, Davies J. 1974. Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins. Proc Nat Acad Sci USA. 71:30–34.
- Desjardins AE, Plattner RD. 2000. Fumonisin B1-nonproducing strains of Fusarium verticillioides cause maize (Zea mays) ear infection and ear rot. J Agric Food Chem. 48:5773–5780.
- Edwards J, Auer D, de Alwis SK, Summerell B, Aoki T, Proctor RH, Busman M, O’Donnell K. 2016. Fusarium agapanthi sp. nov., a novel bikaverin and fusarubin-producing leaf and stem spot pathogen of Agapanthus praecox (African lily) from Australia and Italy. Mycologia 108:981–992.
- Fries EM. 1821. Systema mycologicum. Vol 1. Lundae, Ex Officina Berlingiana. 520 pp.
- Fuchs E, Binder EM, Heidler D, Krska R. 2002. Structural characterization of metabolites after the microbial degradation of type A trichothecenes by the bacterial strain BBSH 797. Food Addit Contam. 19:379–386.
- Gachango E, Hanson LE, Rojas A, Hao JJ, Kirk WW. 2012. Fusarium spp. causing dry rot of seed potato tubers in Michigan and their sensitivity to fungicides. Plant Dis. 96:1767–1774.
- Gale LR, Harrison SA, Ward TJ,, Milus EA, Gale SW, Kistler HC. 2011. Nivalenol-type populations of Fusarium graminearum O’Donnell Kand F. asiaticum are prevalent on wheat in southern Louisiana. Phytopathology 101:124–134.
- Gale LR, Ward TJ, Balmas V, Kistler HC. 2007. Population subdivision of Fusarium graminearum sensu stricto in the upper Midwestern United States. Phytopathology 97:1434–1439.
- Geiser DM, Aoki T, Bacon CW, Baker SE, Bhattacharyya MK, Brandt ME, Brown DW, Burgess LW, Chulze S, Coleman JJ, et al. 2013. One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 103:400–408.
- Geiser DM, Jimenez-Gasco MD, Kang SC, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K. 2004. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 110:473–479.
- Gräfenhan T, Johnston PR, Vaughan MM, McCormick SP, Proctor RH, Busman M, Ward TJ, O’Donnell K. 2016. Fusarium praegraminearum sp. nov., a novel nivalenol mycotoxin-producing pathogen from New Zealand can induce head blight on wheat. Mycologia 108:1229–1239.
- Grudzinska-Sterno M, Yuen J, Stenlid J, Djurle A. 2016. Fungal communities in organically grown winter wheat affected by plant organ and development stage. Eur J Plant Pathol. 146:401–417.
- Guan S, He J, Young JC, Zhu H, Li X-Z, Ji C, Zhou T. 2009. Transformation of trichothecene mycotoxins by microorganisms from fish digesta. Aquaculture 290:290–295.
- Hansen FT, Gardiner DM, Lysoe E, Fuertes PR, Tudzynski B, Wiemann P, Sondergaard TE, Giese H, Brodersen DE, Sorensen JL. 2015. An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium. Fungal Genet Biol. 75:20–29.
- He JW, Yang R, Zhou T, Boland GJ, Scott PM, Bondy GS. 2015. An epimer of deoxynivalenol: purification and structure identification of 3-epi-deoxynivalenol. Food Addit Contam. 32:1523–1530.
- Hertz M, Jensen IR, Jensen LO, Thomsen SN, Winde J, Dueholm MS, Sorensen LH, Wollenberg RD, Sorensen HO, Sondergaard TE, Sorensen JL. 2016. The fungal community changes over time in developing wheat heads. Int J Food Microbiol. 222:30–39.
- Ikunaga Y, Sato I, Grond S, Numaziri N, Yoshida S, Yamaya H, Hiradate S, Hasegawa M, Toshima H, Koitabashi M, et al. 2011. Nocardioides sp. strain WSN05-2, isolated from a wheat field, degrades deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol. Appl Microbiol Biotechnol. 89:419–427.
- Karlsson I, Friberg H, Steinberg C, Persson P. 2014. Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS ONE. 9:e111786.
- Kazan K, Gardiner DM, Manners JM. 2012. On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol Plant Pathol. 13:399–413.
- Kelly AC, Clear RM, O’Donnell K, McCormick SP, Turkington TK, Tekauz A, Gilbert J, Kistler HC, Busman M, Ward TJ. 2015. Diversity of Fusarium head blight populations and trichothecene toxin types reveals regional differences in pathogen composition and temporal dynamics. Fungal Genet Biol. 82:22–31.
- Kelly AC, Proctor RH, Belzile F, Chulze SN, Clear RM, Cowger C, Elmer W, Lee T, Obanor F, Waalwijk C, Ward TJ. 2016. The geographic distribution and complex evolutionary history of the NX-2 trichothecene chemotype from Fusarium graminearum. Fungal Genet Biol. 95:39–48.
- Kim H-S, Proctor RH, Brown DW. 2017. Comparative genomic analyses of secondary metabolite biosynthetic gene clusters in 207 isolates of Fusarium. In: 29th Fungal Genetics Conference. Genetics Society of America, Pacific Grove, CA. p. 170.
- Kim YT, Lee YR, Jin J, Han KH, Kim H, Kim JC, Lee T, Yun SH, Lee YW. 2005. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae. Mol Microbiol. 58:1102–1113.
- Kimura M, Kaneko I, Komiyama M, Takatsuki A, Koshino H, Yoneyama K, Yamaguchi I. 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins: cloning and characterization of Tri101. J Biol Chem. 273:1654–1661.
- Kimura M, Tokai T, O’Donnell K, Ward TJ, Fujimura M, Hamamoto H, Shibata T, Yamaguchi I. 2003. The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett. 539:105–110.
- Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M. 2007. Molecular and genetic studies of Fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol Biochem. 71:2105–2123.
- Kowalska K, Habrowska-Górczyńska DE, Piastowska-Ciesielska AW. 2016. Zearalenone as an endocrine disruptor in humans. Environ Toxicol Pharmacol. 48:141–149.
- Laurence MH, Summerell BA, Burgess LW, Liew ECY. 2011. Fusarium burgessii sp. nov. representing a novel lineage in the genus Fusarium. Fungal Div. 49:101–112.
- Lee T, Han Y-K, Kim K-H, Yun S-H, Lee Y-W. 2002. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl Environ Microbiol. 68:2148–2154.
- Lemmens M, Scholz U, Berthiller F, Dall’Asta C, Koutnik A, Schuhmacher R, Adam G, Buerstmayr H, Mesterhazy A, Krska R, Ruckenbauer P. 2005. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol Plant-Microbe Interact. 18:1318–1324.
- Li X, Shin S, Heinen S, Dill-Macky R, Berthiller F, Nersesian N, Clemente T, McCormick SP, Muehlbauer GJ. 2015. Transgenic wheat expressing a barley UDP-glucosyltransferase detoxifies deoxynivalenol and provides high levels of resistance to Fusarium graminearum. Mol Plant-Microbe Interact. 28:1237–1246.
- Li XZ, Zhu C, de Lange CF, Zhou T, He J, Yu H, Gong J, Young JC. 2011. Efficacy of detoxification of deoxynivalenol-contaminated corn by Bacillus sp. LS100 in reducing the adverse effects of the mycotoxin on swine growth performance. Food Addit Contam. 28:894–901.
- Liang J, Lofgren L, Ma Z, Ward TJ, Kistler HC. 2015. Population subdivision of Fusarium graminearum from barley and wheat in the upper Midwestern United States at the turn of the century. Phytopathology 105:1466–1474.
- Liang JM, Xayamongkhon H, Broz K, Dong Y, McCormick SP, Abramova S, Ward TJ, Ma ZH, Kistler HC. 2014. Temporal dynamics and population genetic structure of Fusarium graminearum in the upper Midwestern United States. Fungal Genet Biol. 73:83–92.
- Lin FY, Lu QX, Xu JH, Shi JR. 2008. Cloning and expression analysis of two salt and Fusarium graminearum stress associated UDP-glucosyltransferases genes in wheat [article in Chinese]. Hereditas 30:1608–1614.
- Link HF. 1809. Observationes in ordines plantarum naturales. Dissertatio I. Mag Ges naturf Freunde, Berlin. 3:10(Tab I, Fig 10).
- Lysøe E, Harris LJ, Walkowiak S, Subramaniam R, Divon HH, Riiser ES, Llorens C, Gabaldón T, Kistler HC, Jonkers W, et al. 2014. The genome of the generalist plant pathogen Fusarium avenaceum is enriched with genes involved in redox, signaling and secondary metabolism. PLoS ONE. 9:e112703.
- Ma LJ, Geiser DM, Proctor RH, Rooney AP, O’Donnell K, Trail F, Gardiner DM, Manners JM, Kazan K. 2013. Fusarium pathogenomics. Annu Rev Microbiol. 67:399–416.
- Maragos CM, Kurtzman C, Busman M, Price N, McCormick S. 2013. Development and evaluation of monoclonal antibodies for the glucoside of T-2 toxin (T2-Glc). Toxins 5:1299–1313.
- McCormick SP. 2013. Microbial detoxification of mycotoxins. J Chem Ecol. 39:907–918.
- McCormick SP, Alexander NJ, Trapp SE, Hohn TM. 1999. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl Environ Microbiol. 65:5252–5256.
- McCormick SP, Harris LJ, Alexander NJ, Ouellet T, Saparno A, Allard S, Desjardins AE. 2004. Tri1 in Fusarium graminearum encodes a P450 oxygenase. Appl Environ Microbiol. 70:2044–2051.
- McCormick SP, Kato T, Maragos CM, Busman M, Lattanzio VM, Galaverna G, Dall-Asta C, Crich D, Price NP, Kurtzman CP. 2015. Anomericity of T-2 toxin-glucoside: masked mycotoxin in cereal crops. J Agric Food Chem. 63:731–738.
- McCormick SP, Price NP, Kurtzman CP. 2012. Glucosylation and other biotransformations of T-2 toxin by yeasts of the Trichomonascus clade. Appl Environ Microbiol. 78:8694–8702.
- Meek IB, Peplow AW, Ake J, Charles, Phillips TD, Beremand MN. 2003. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl Environ Microbiol. 69:1607–1613.
- Michielse CB, Rep M. 2009. Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol. 10:311–324.
- Munkvold GP. 2017. Fusarium species and their associated mycotoxins. In Mycotoxigenic Fungi: Methods and Protocols. Edited by A. Moretti and A. Susca. Springer New York, New York, NY. pp. 51–106.
- Nganje WE, Bangsund DA, Leistritz FL, Wilson WW, Tiapo NM. 2004. Regional economic impacts of Fusarium head blight in wheat and barley. Rev Agric Econ. 26:332–347.
- Nicolaisen M, Justesen AF, Knorr K, Wang J, Pinnschmidt HO. 2014. Fungal communities in wheat grain show significant co-existence patterns among species. Fungal Ecol. 11:145–153.
- Niehaus E-M, Janevska S, von Bargen KW, Sieber CMK, Harrer H, Humpf H-U, Tudzynski B. 2014. Apicidin F: characterization and genetic manipulation of a new secondary metabolite gene cluster in the rice pathogen Fusarium fujikuroi. PLoS ONE. 9:e103336.
- Niehaus E-M, Kleigrewe K, Wiemann P, Studt L, Sieber Christian MK, Connolly Lanelle R, Freitag M, Güldener U, Tudzynski B, Humpf H-U. 2013. Genetic manipulation of the Fusarium fujikuroi fusarin gene cluster yields insight into the complex regulation and fusarin biosynthetic pathway. Chem Biol. 20:1055–1066.
- Niehaus EM, Munsterkotter M, Proctor RH, Brown DW, Sharon A, Idan Y, Oren-Young L, Sieber CM, Novak O, Pencik A, et al. 2016. Comparative “omics” of the Fusarium fujikuroi species complex highlights differences in genetic potential and metabolite synthesis. Genome Biol Evol. 8:3574–3599.
- O’Donnell K, Rooney AP, Proctor RH, Brown DW, McCormick SP, Ward TJ, Frandsen RJ, Lysøe E, Rehner SA, Aoki T, et al. 2013. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet Biol. 52:20–31.
- O’Donnell K, Ward TJ, Robert VARG, Crous PW, Geiser DM, Kang S. 2015. DNA sequence-based identification of Fusarium: current status and future directions. Phytoparasitica 43:583–595.
- Palumbo JD, O’Keeffe TL, Abbas HK. 2008. Microbial interactions with mycotoxigenic fungi and mycotoxins. Toxin Rev. 27:261–285.
- Peplow AW, Meek IB, Wiles MC, Phillips TD, Beremand MN. 2003. Tri16 is required for esterification of position C-8 during trichothecene mycotoxin production by Fusarium sporotrichioides. Appl Environ Microbiol. 69:5935–5940.
- Pestka J. 2010. Toxicological mechanisms and potential health effects of deoxynivalenol and nivalenol. World Mycotoxin J. 3:323–347.
- Proctor RH, Hohn TM, McCormick SP. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant-Microbe Interact. 8:593–601.
- Proctor RH, McCormick SP, Alexander NJ, Desjardins AE. 2009. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol Microbiol. 74:1128–1142.
- Proctor RH, Van Hove F, Susca A, Stea G, Busman M, van der Lee T, Waalwijk C, Moretti A, Ward TJ. 2013. Birth, death and horizontal transfer of the fumonisin biosynthetic gene cluster during the evolutionary diversification of Fusarium. Mol Microbiol. 90:290–306.
- Reynolds H, Slot JC, Divon HH, Lysøe E, Proctor RH, Brown DW. 2017. Differential retention of gene functions in a secondary metabolite cluster. Mol Biol Evol. 34:2002–2015.
- Sarver BA, Ward TJ, Gale LR, Broz K, Kistler HC, Aoki T, Nicholson P, Carter J, O’Donnell K. 2011. Novel Fusarium head blight pathogens from Nepal and Louisiana revealed by multilocus genealogical concordance. Fungal Genet Biol. 48:1096–1107.
- Sato I, Ito M, Ishizaka M, Ikunaga Y, Sato Y, Yoshida S, Koitabashi M, Tsushima S. 2012. Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. FEMS Microbiol Lett. 327:110–117.
- Schmale DG, Wood-Jones AK, Cowger C, Bergstrom GC, Arellano C. 2011. Trichothecene genotypes of Gibberella zeae from winter wheat fields in the eastern USA. Plant Pathol. 60:909–917.
- Semeiks J, Borek D, Otwinowski Z, Grishin NV. 2014. Comparative genome sequencing reveals chemotype-specific gene clusters in the toxigenic black mold Stachybotrys. BMC Genom. 15:590.
- Sieber CM, Lee W, Wong P, Munsterkotter M, Mewes HW, Schmeitzl C, Varga E, Berthiller F, Adam G, Guldener U. 2014. The Fusarium graminearum genome reveals more secondary metabolite gene clusters and hints of horizontal gene transfer. PLoS ONE. 9:e110311.
- Stockmann-Juvala H, Savolainen K. 2008. A review of the toxic effects and mechanisms of action of fumonisin B1. Human Exp Toxicol. 27:799–809.
- Studt L, Troncoso C, Gong F, Hedden P, Toomajian C, Leslie JF, Humpf HU, Rojas MC, Tudzynski B. 2012. Segregation of secondary metabolite biosynthesis in hybrids of Fusarium fujikuroi and Fusarium proliferatum. Fungal Genet Biol. 49:567–577.
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 31:21–32.
- Uhlig S, Busman M, Shane DS, Rønning H, Rise F, Proctor R. 2012. Identification of early fumonisin biosynthetic intermediates by inactivation of the FUM6 gene in Fusarium verticillioides. J Agric Food Chem. 60:10293–10301.
- Uhlig S, Petersen D, Flaoyen A, Wilkins A. 2005. 2-Amino-14,16-dimethyloctadecan-3-ol, a new sphingosine analogue toxin in the fungal genus Fusarium. Toxicon 46:513–522.
- Vanheule A, Audenaert K, Warris S, van de Geest H, Schijlen E, Hofte M, De Saeger S, Haesaert G, Waalwijk C, van der Lee T. 2016. Living apart together: crosstalk between the core and supernumerary genomes in a fungal plant pathogen. BMC Genom. 17:670.
- Vanhoutte I, Audenaert K, De Gelder L. 2016. Biodegradation of mycotoxins: tales from known and unexplored worlds. Frontier Microbiol. 7:561.
- Varga E, Wiesenberger G, Hametner C, Ward TJ, Dong Y, Schofbeck D, McCormick S, Broz K, Stuckler R, Schuhmacher R, et al. 2015. New tricks of an old enemy: isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ Microbiol. 17:2588–2600.
- Vaughan MM, Backhouse D, Ponte EMD. 2016a. Climate change impacts on the ecology of Fusarium graminearum species complex and susceptibility of wheat to Fusarium head blight: a review. World Mycotoxin J. 9:685–700.
- Vaughan MM, Huffaker A, Schmelz EA, Dafoe NJ, Christensen S, Sims J, Martins VF, Swerbilow J, Romero M, Alborn HT, et al. 2014. Effects of elevated [CO2] on maize defence against mycotoxigenic Fusarium verticillioides. Plant Cell Environ. 37:2691–2706.
- Vaughan MM, Huffaker A, Schmelz EA, Dafoe NJ, Christensen SA, McAuslane HJ, Alborn HT, Allen LH, Teal PE. 2016b. Interactive effects of elevated [CO2] and drought on the maize phytochemical defense response against mycotoxigenic Fusarium verticillioides. PLoS ONE 11:e0159270.
- Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K. 2002. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Nat Acad Sci USA. 99:9278–9283.
- Ward TJ, Clear RM, Rooney AP, O’Donnell K, Gaba D, Patrick S, Starkey DE, Gilbert J, Geiser DM, Nowicki TW. 2008. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet Biol. 45:473–484.
- Wetterhorn KM, Newmister SA, Caniza RK, Busman M, McCormick SP, Berthiller F, Adam G, Rayment I. 2016. Crystal structure of Os79 (Os04g0206600) from Oryza sativa: a UDP-glucosyltransferase involved in the detoxification of deoxynivalenol. Biochemistry 55:6175–6186.
- Wiemann P, Sieber CMK, von Bargen KW, Studt L, Niehaus E-M, Espino JJ, Huß K, Michielse CB, Albermann S, Wagner D, et al. 2013. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathogens 9:e1003475.
- Windels CE. 2000. Economic and social impacts of Fusarium head blight: changing farms and rural communities in the Northern Great Plains. Phytopathology 90:17–21.
- Wu F, Groopman JD, Pestka JJ. 2014. Public health impacts of foodborne mycotoxins. Annu Rev Food Sci Technol. 5:351–372.
- Yoshida S, Ohba A, Liang YM, Koitabashi M, Tsushima S. 2012. Specificity of Pseudomonas isolates on healthy and Fusarium head blight-infected spikelets of wheat heads. Microb Ecol. 64:214–225.
- Young CA, Kinkel L. 2017. Welcome to Phytobiomes. Phytobiomes J. 1:3–4.
- Young JC, Zhou T, Yu H, Zhu H, Gong J. 2007. Degradation of trichothecene mycotoxins by chicken intestinal microbes. Food Chem Toxicol. 45:136–143.
- Yu H, Zhou T, Gong J, Young C, Su X, Li XZ, Zhu H, Tsao R, Yang R. 2010. Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. 10:182.
- Zhou T, He J, Gong J. 2008. Microbial transformation of trichothecene mycotoxins. World Mycotoxin J. 1:23–30.
- Zhou X, O’Donnell K, Aoki T, Smith JA, Kasson MT, Cao ZM. 2016. Two novel Fusarium species that cause canker disease of prickly ash (Zanthoxylum bungeanum) in northern China form a novel clade with Fusarium torreyae. Mycologia 108:668–681.
