 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The recycling of plastics such as polyurethane (PU) is a current challenge. The continuously increasing production volume and additionally growing end-of-life streams increase the urgency for solutions. Conventional recycling methods such as mechanical and chemical recycling are only economically and/or ecologically suitable to a limited extent. The three-step approach consisting of enzymatic PU decomposition, separation of amines (4,4’-methylenedianiline (MDA), isophoronediamine (IPDA), 2,4-toluenediamine (TDA)) and fermentation of the residual stream is a promising recycling concept. In this study, extraction methods for the separation of MDA and IPDA from an aqueous solution at neutral pH are developed. In addition, the influence of relevant PU hydrolysate components on the amine extraction is investigated. The results show that MDA can be efficiently separated using solvent extraction of 1-octanol. For IPDA separation, a reactive extraction with oleic acid as reactant is developed. The application of these two extraction methods to TDA shows extraction efficiencies of 52% to 86%. The other PU hydrolysate components adipic acid and selected salts have only a minor influence on the extraction efficiency. The diols ethylene glycol and 1,4-butanediol influence the equilibrium pH of IPDA extraction, raising it to higher values. For MDA, no influence of other PU hydrolysate components on the extraction efficiency can be observed. Since amines can have an inhibitory effect on microorganisms, toxicity experiments were carried out to determine the tolerable residual concentration of amines in the raffinate to avoid fermentation inhibition. Growth experiments with Pseudomonas putida KT240 show that MDA has an inhibitory effect at concentrations near the solubility limit, whereas IPDA does not affect growth.
Introduction
With the “Green Deal”, the European Union (EU) is following the goal of achieving climate neutrality in Europe by the year 2050.[Citation1] An important step towards this goal is a comprehensive circular economy, whereby the recycling of polymers poses a particular challenge. This challenge is reflected by polymers such as polyurethane (PU), which shows a high landfill rate of almost 50% in the EU.[Citation2] There are different approaches to tackle this challenge, such as mechanical recycling, or chemical recycling by chemolysis or pyrolysis.[Citation3–8] A promising three-step approach to recycle PU is based on biocatalytic decomposition, purification and subsequent fermentation of the residues,[Citation9,Citation10] yielding different valuable base building blocks.[Citation11]
First, the PU is depolymerized by enzymatic hydrolysis via oxidoreductases or hydrolases. Typical degradation products resulting from the cleavage are 4,4’-methylenedianiline (MDA), 2,4-toluenediamine (TDA), isophoronediamine (IPDA), butane-1,4-diol (BDO), ethane-1,2-diol (EG) and adipic acid (AA).[Citation12,Citation13] The separation of these components by extraction into amines and the remaining lower-value monomers is a key step for recycling. The separated aromatic amines (MDA, TDA, IPDA) can be reused as base building blocks for the production of virgin PU.[Citation14] The remaining lower-value monomers can be upcycled into high-value chemicals such as rhamnolipids via microbiological fermentation using a Pseudomonas strain. Rhamnolipids are used, for example, as biosurfactant in detergents. Since amines can have an inhibitory effect on the Pseudomonas strain, the tolerable residual concentration of amines is fundamental information to develop the extraction.[Citation10]
This was already shown during the microbial conversion of a TDA-based model PU hydrolysate, in which the toxicity of TDA on microorganisms was proven. To selectively recover the TDA from the aqueous hydrolysate, a reactive extraction technique using di-(2-ethylhexyl) phosphoric acid (D2EHPA) as reactant was applied. This approach reduced the TDA concentration so that the inhibitory effect of TDA on the applied Pseudomonas strain could be significantly lowered. However, Utomo’s group has found that extraction creates a new inhibition. The reactant D2EHPA is partially soluble in water and itself has an inhibitory effect on fermentation. To mitigate this effect, extraction was performed at a low pH, where the cross-solubility of D2EHPA is reduced. Since fermentation occurs at a neural pH, a pH adjustment between extraction and fermentation is required.[Citation10]
In the three-step approach for PU recycling, enzymes are used in the first step and microorganisms in the last step, which requires a neutral pH. Since the extraction takes place between these two stages, this work aims to develop an extraction of aromatic amines at neutral pH. More specifically, this paper focuses on the recovery of MDA and IPDA from hydrolysates by solvent extraction and reactive extraction and thus addresses further relevant aromatic amines derived from PU hydrolysates.
In addition to the extraction technique investigations, the inhibitory effect of MDA and IPDA on a Pseudomonas strain was investigated in toxicity studies. The objective of these studies is to determine acceptable residual amine concentrations to provide further insight into the requirements for successful integration of extraction into the three-stage recycling process presented above.
Materials and methods
Chemicals
Isophoronediamine 99+ % (IPDA), adipic acid 99% (AA) and 2,4-toluenediamine 98% (TDA) were purchased from Acros Organics (Geel, Belgium). 4,4′-methylenedianiline ≥97% (MDA), oleic acid FCC grade (OLA) and adipic acid for the toxicity experiment were purchased from Sigma-Aldrich (St. Louis, MO, USA). 1,4butanediol ≥99% (BDO) was purchased from Carl Roth (Karlsruhe, Germany) and 1,2-ethanediol 99% and 1-octanol 99% were purchased from Thermo Fischer (Kandel, Germany). All other chemicals came from the companies mentioned above or from Merck (Darmstadt, Germany) and VWR (Leuven, Belgium).
Microbiological toxicity experiment for MDA and IPDA
For quantitative microbiology toxicity experiments, P. putida KT2440 and the engineered adipic acid-metabolizing KT2440ge ΔPpaaF-paaYX::P14 g ΔpsrA were cultivated in mineral salts medium (MSM) with a buffer concentration three times higher compared to Wierckx et al. to prevent pH shift after degradation of acidic substrates. (11.64 g/L K2HPO4, 4.89 g/L NaH2PO4).[Citation15,Citation16] For the cultivation with AA, a 300 mM AA stock solution was dissolved 1:10 in MSM to reach a final concentration of 30 mM. For the different MDA and IPDA concentrations, 10-fold stocks were prepared starting from 5 mM and then diluted 1:10 with the medium. Due to the low solubility of MDA which is 1.01 g/L, a maximum concentration of 0.85 g/L (4.3 mM) was chosen for MDA experiments.[Citation17] For online growth detection a Growth Profiler® 960 (Enzyscreen, Heemstede, The Netherlands) was used. This device analyses cultures in microtiter plates with transparent bottoms by image analysis. Pre-cultures containing 2 mL MSM with 20 mM glucose in 14 mL culture tubes (Greiner bio-one, Frickenhausen, Germany) were cultivated in a Multitron shaker (INFORS, Bottmingen, Switzerland) with a 220 rpm shaking speed. Main cultures in 96-well plates with 200 µL volume, using MSM with several concentrations of different carbon sources as indicated, were incubated at 30°C, 225 rpm shaking speed with an amplitude of 50 mm in the Growth Profiler® in biological triplicates. Pictures were taken every 30 minutes and analysed using the Growth Profiler Control software V2_0_0. The resulting green-values (G-values, based on green pixel counts) correlate with the optical density of a cell culture. These G-values were converted into optical densities at a wavelength of λ = 600 nm (OD600) equivalents using a calibration curve for P. putida.[Citation18]
Solvent and reactive extraction experiments
As was already shown for TDA, also MDA and IPDA (see ) are proton acceptors and show pH dependent dissociation behavior.
Figure 1. Chemical structure of 4,4’-methylenedianiline (MDA), 2,4-toluenediamine (TDA) and isophoronediamine (IPDA).
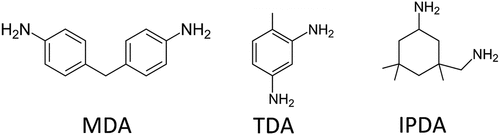
The two functional amine groups of the investigated substances can have three different dissociation states, resulting in pH-dependent extraction behavior.[Citation19] A detailed discussion of the pH-dependent extraction behavior is given in the section Extraction of diamines. For this discussion and comparison, the extraction efficiency is introduced, which is defined as
the ratio of the mass of component i of the organic phase in the equilibrium state , to the mass of component i of the aqueous phase in the initial state
. The mass of the components in the organic phase, is calculated based on the overall mass balance and the components mass in the aqueous phase. The mass balance is given in the following.
With the assumption of constant volume of the aqueous phase in the initial and equilibrium state, and a concentration measurement of component i in the aqueous phase at the initial state and equilibrium state by means of HPLC, the extraction efficiency E can be calculated by
Separation of diamines were studied in single-stage solvent extraction and reactive extraction experiments. For solvent extraction, 1-octanol was used as solvent. For reactive extraction, OLA as reactant diluted in 1-octanol was used as organic phase. 1-octanol was chosen as diluent since it has been reported in the literature as a suitable solvent for bioprocesses and has already been successfully used in biotechnological processes in a reactive in situ extraction.[Citation20,Citation21] In addition, it is non-toxic, only slightly soluble in water and can be produced biotechnologically from renewable raw materials.[Citation22] If not specified otherwise, the concentration of the reactive agent was 0.1 M. For the extraction, 1 mL of the corresponding aqueous diamine solution with a concentration of 0.5 g/L, were mixed with 1 mL solvent in 2 mL tubes. This concentration corresponds to 2.5 mM for MDA, 2.9 mM for IPDA and 4.1 mM for TDA. The water used was distilled in a MonoDest3000 (Lenz Glas Instrumente, Wertheim, Germany). An orbital Lab-Shaker LS-W (Kuehner, Birsfelden, Germany) at 150 rpm for at least 14 h realized mixing of organic and aqueous phase. After this time, it was assumed that the thermodynamic equilibrium state is reached. For phase separation, the tubes were centrifuged at 10,000 rpm for one minute with an IKA G-L centrifuge (IKA, Staufen, Germany). Before and after extraction, the amine concentration of the aqueous phase was determined via HPLC. The error of the extraction experiments is determined with the coefficient of variation based on the concentration measurement via HPLC of all samples of each experiment before extraction. This is defined as the ratio of the standard deviation
to the arithmetic mean
:
The coefficient of variation is given in each figure caption.
The pH value of the aqueous phase was measured in the equilibrium state after the extraction with a SevenCompact pH/Ionmeter (Mettler Toledo, Gießen, Germany). All experiments were conducted at ambient temperature 295 K ± 1 K. Sulfuric acid, hydrochloric acid, citric acid, sulfates and phosphates were used to adjust the pH and as buffers. Table S1 in the supporting information lists the substances and concentrations used for each experiment. In addition, the pH adjusting agents used are mentioned under each figure.
Furthermore, the influence of diols (EG, BDO), carboxylic acid AA and salts (NaCl, phosphate, sulphate) were investigated at concentrations up to 500 mM.
HPLC analytics for diamine quantification
For the determination of the amine concentration of the aqueous phase, an Agilent 1200 HPLC system (Agilent, Santa Clara, USA) equipped with a Diode-Array Detection (DAD) was used. The method for the detection of MDA and TDA is shown in .
Table 1. HPLC method for MDA and TDA analysis.
For the concentration determination via HPLC of IPDA the method of Doeker et al.[Citation23] was used, for diols and carboxylic acid the method of Kocks et al.[Citation24]
Results & discussion
Microbiological toxicity experiment for MDA and IPDA
Experiments were carried out to determine the influence of MDA and IPDA on growth of P. putida KT2440 on glucose or adipate as sole carbon source. Both P. puitda KT2440 wild-type and a engineered KT2440 strain optimized for the degradation of adipate were used for this purpose.[Citation16] The growth rates are shown in . In both strains, the highest concentration of MDA (4.3 mM) resulted in complete growth inhibition (). At the concentration of 0.5 mM MDA no impaired growth was observed compared to the negative control without MDA when glucose was used as sole carbon source. If adipate as common plastic monomer is used as the sole carbon source, growth was slightly impaired at 0.5 mM MDA. Concentrations below 0.05 mM showed no significant effect on the growth of the two P. putida strains cultured either glucose or adipate.
Figure 2. Characterization of bacterial growth under different MDA concentrations. Two strains of P. putida (A = wild-type KT2440, B = reverse engineered KT2440ge ΔPpaaF-paaYX:P14 g ΔpsrA) were grown in MSM medium with either 20 mM glucose or 30 mM adipate supplemented with different MDA concentrations (0 mM, 0.0005 mM, 0.005 mM, 0.05 mM, 0.5 mM, 4.3 mM MDA). OD600 equivalents (OD600 eq.) were derived from green-values obtained from the Growth Profiler using a calibration curve. Symbols show every 2nd data point. Error bars indicate the standard error of the mean (n = 3).
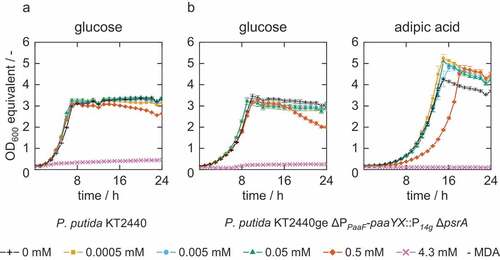
Table 2. Bacterial growth rates under different MDA concentrations for the two strains of P. putida (A = wild-type KT2440, B = reverse engineered KT2440ge ΔPpaaF-paaYX::P14 g ΔpsrA).
IPDA was clearly less toxic than MDA. None of the tested concentrations (0 mM − 5.9 mM IPDA) had any negative effects on growth, regardless of which carbon source was used and whether it was the wild-type or the reverse-engineered strain. The results can be found in the supporting information in Figure S1. As previously mentioned, the toxicity of TDA has already been proven by Utomo et al.[Citation10]
Extraction of diamines
Solvent extraction of MDA and IPDA
shows the pH dependent extraction efficiency of MDA and IPDA via solvent extraction. For both diamines, a strong dependency of extraction efficiency on pH was observed. For MDA, no extraction was observed at pH values below 3. For pH between 3 and 5, a continuous increase in MDA extraction was observed, and very high extraction efficiencies of up to E = 97.6% (logP = 1.61) were achieved from pH higher than approximately 6. IPDA showed a similar behaviour, only with a shift to much higher pH values. For IPDA below pH 7 no extraction took place, between pH 7 and 10 the extraction efficiency increased continuously, and above pH 13 a maximum was reached with a extraction efficiency of E = 98.1% (logP = 1.71).
Figure 3. Extraction efficiency of solvent extraction of 2.5 mM MDA (aq) and 2.9 mM IPDA (aq) from an aqueous solution with 1-octanol. The pH value was determined at the equilibrium state. Adjustment of the pH with sulphuric acid and sodium hydroxide. νMDA=1.5%, νIPDA=2.4%.
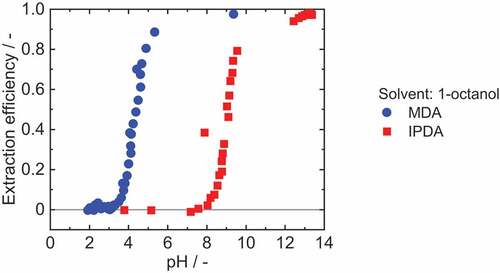
This difference in the pH dependency can be explained by the dissociation behaviour of MDA and IPDA which is defined by the pKa value. In the literature, the pKa1 values are reported with 5.3 for MDA and 10.7 for IPDA.[Citation17,Citation25] At pH below the pKa, the protonated species is predominant, while at pH above the pKa the unprotonated mainly occurs. The protonated and thus positively charged species cannot be extracted. Consequently, the extraction efficiency is decreasing below the pKa and increasing above the pKa, as shown in .
Therefore, MDA can be extracted from an aqueous solution via solvent extraction at neutral pH, whereas IPDA requires a pH lager than 10.7 for an efficient extraction. This means that further pH adjustment would be required if IPDA separation were performed by solvent extraction, since enzymatic hydrolysis and fermentation are typically performed at near-neutral pH.[Citation23,Citation26] This pH adjustment would lead to an increased salt load, which results in larger waste streams and higher costs. Thus, an alternative separation technique such as reactive extraction is appropriate for efficient separation of IPDA.
Reactive extraction of IPDA
To achieve high extraction efficiencies for IPDA at neutral pH, reactive extraction was applied. OLA can form a reactive complex with the protonated amine, which has a high affinity for the organic phase. The formation of the complex is strongly pH dependent and thus also leads to a pH dependence of the extraction.[Citation23] First, the influence of the OLA concentration, on the extraction efficiencywas investigated. The results of these extraction experiments for four different OLA concentrations (0.1 M to 1 M) and without OLA are given in . Variation in OLA concentration resulted in a shift in the extraction curves, with higher OLA concentrations leading to a lower equilibrium pH. Thus, via variation of the OLA concentration, the pH operating window of the extraction can be adjusted. Based on the results of the literature, we assume that the reactive extraction is a liquid cation exchanger. Moreover, we assume a similar mechanism as already described in the work of Doeker and Bednarz.[Citation19,Citation23]
Figure 4. Extraction efficiency for reactive extraction of 2.9 mM IPDA (aq) with 1-octanol as diluent and different OLA concentrations as reactant. Adjustment of the pH with citric acid, sulphuric acid and sodium hydroxide. ν1.00 M=3.2%, ν0.50 M=2.4%, ν0.25 M=2.9%, ν0.10 M=4.3%.
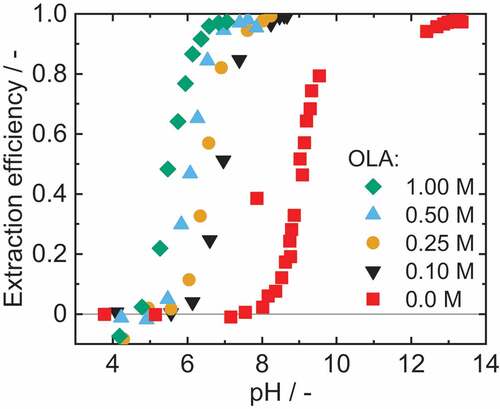
Without OLA, a pure solvent extraction occurs, in which high extraction efficiencies can only be achieved at pH greater 12. Whereas reactive extraction allows high extraction efficiencies even at neutral pH.
Influence of solutes from PU hydrolysis reactive extraction of IPDA
Besides the diamines, the PU hydrolysate is composed of a variety of components depending on PU composition and additives.[Citation13] These components can influence the selectivity of the reactive extraction, and thus affect the overall efficiency of the process. To characterize this influence, representative components were identified and added to the extraction experiments. The identified components are diols (EG, BDO), adipic acids (AA) and salts (sodium chloride, sodium phosphates). For all subsequent experiments, the results from the previous reactive extraction experiments were used as a reference, allowing to show differences in diamine extraction.
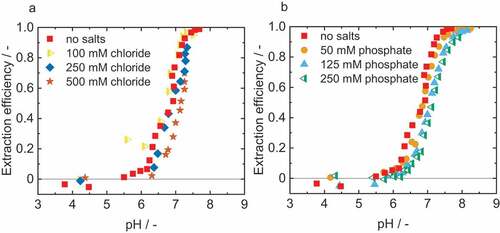
depicts the extraction efficiency for the reactive extraction of IPDA with different relevant salts. For both chlorides and phosphates, a shift of the extraction curves towards higher pH can be observed. At low salt concentrations, there are only slight deviations from the experiments without salt addition with a pH difference of mostly less than 0.1. With increasing salt concentration, this difference increases up to a pH difference of 0.7. A possibility to compensate the pH shift would be to increase the OLA concentration. However, a reduction of the maximum extraction efficiency could not be observed.
Figure 5. Extraction efficiency for reactive extraction of 2.9 mM IPDA (aq) with (a) chlorides and (b) phosphates. 1-octanol was used as diluent and 0.1 M oleic acid as reactant. Adjustment of the pH with sulphuric acid and sodium hydroxide. νno salt=2.0%, ν100 mM chloride=1.5%, ν250 mM chloride=1.4%, ν500 mM chloride=1.2%, ν50 mM phosphate=1.5%, ν125 mM phosphate=1.6%,ν250 mM phosphate=1.1%.
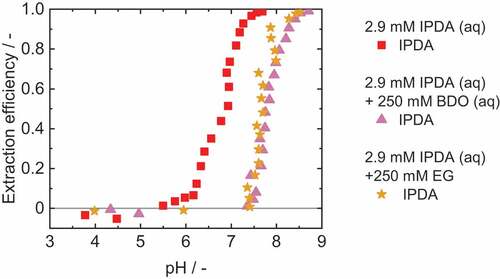
The influence of BDO and EG on the reactive extraction of IPDA is shown in . With both BDO and EG, the extraction efficiency is shifted to higher pH values. The pH difference for both diols compared to the reference experiment is about 1. However, the maximum extraction efficiency is not affected by the diols.
Figure 6. Extraction efficiency of IPDA with 1-octanol as diluent and 0.1 M OLA as reactant. Adjustment of the pH with sulphuric acid and sodium hydroxide. νIPDA+BDO=0.7%, νIPDA+EG=0.8%.
In addition to the amine extraction, the co-extraction of the diols was also investigated. The results are shown in the supporting information in figure S3 and show that both EG and BDO was extracted with a constant extraction efficiency of approx. E = 4% and E = 12%, respectively, independent of the pH value. The resulting partition coefficient logP corresponds to the partition coefficients from the literature with logPEG = −1.4 and logPBDO = −0.9.[Citation17,Citation27] This co-extraction leads to a reduction of the diol concentration in the raffinate and is thus not available as a carbon source for fermentation. In addition, it leads to contamination of the extract and impedes further downstream processes in order to reuse the amines for the production of virgin PU.Thus, without further process steps a selective IPDA single-stage extraction is challenging. A possible approach to increase selectivity would be a multi-step extraction.
Furthermore, co-extraction of diols affects the extraction behavior. One explanation is that co-extraction of significant amounts of diols would lead to dilution of the organic phase and lower OLA concentrations, resulting in a shift of the extraction curve to higher pH. However, since the co-extracted amount of diols is small (12% for BDO) and only leads to a decrease in OLA concentration of about 0.28 mM (initially 100 mM), this cannot explain the observed behavior.
A second explanation of the effect could be the influence of the diols on the complex formation and the hydrophobicity of the organic phase, which could lead to a shift in pH. Similar effects have already been observed in the literature for reactive extraction when the diluent was changed.[Citation28] However, this is beyond the scope of this work and will be investigated in another study.
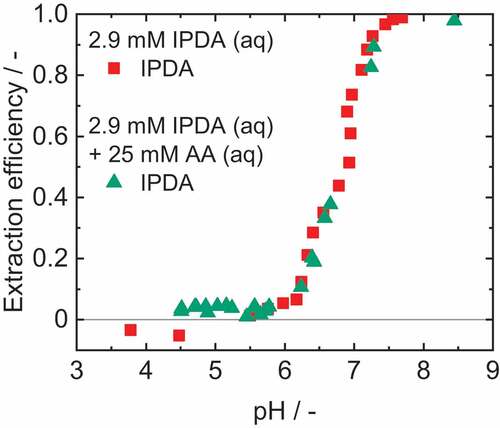
The influence of AA on the reactive extraction is shown in . The data points of the reference experiment and the experiment with AA are superimposed and no influence on the extraction efficiency could be observed.
Figure 7. Extraction efficiency of IPDA with 1-octanol as diluent and 0.1 M OLA as reactant. Adjustment of the pH with chloric acid and sodium hydroxide. νIPDA+AA=1.4%.
Co-extraction was also investigated for AA. The results are shown in the supporting information in Figure S6. Depending on the pH, AA is in different dissociation states, which strongly influences the extraction. At pH below 6, AA is present in protonated state, which was physically extracted to the organic phase. Above pH 6, no protonated species of AA is present in the solution,[Citation29] thus no extraction of AA was observed. This allows selective separation of IPDA at a neutral pH.
For the most part, similar observations regarding the influence of minor components on extraction were made for solvent extraction of MDA as for reactive extraction of IPDA. Therefore, the figures are shown in the supporting information. Figure S2 shows the results for salt influence. Similar to IPDA, the addition of salt results in a small pH shift in extraction efficiency in the range of large pH gradients. The minimum and maximum extraction efficiencies are not affected by the salts investigated. Carboxylic acid also has negligible effect on MDA extraction. The pH shift shown for IPDA caused by the diols cannot be observed for MDA in Figure S3. Also, the co-extraction of the minor components is as previously described for IPDA.
Transfer of the solvent and reactive extraction to TDA
Since MDA was successfully recovered via solvent extraction and the developed reactive extraction showed good results for IPDA separation, both methods were tested for separation of TDA. The results are depicted in .
Figure 8. Extraction efficiency of solvent extraction and reactive extraction of 4.1 mM TDA. Adjustment of the pH with citric acid and sodium hydroxide. ν0 M=1.1%,ν1 M=1.0%,νpure OLA=1.4%.
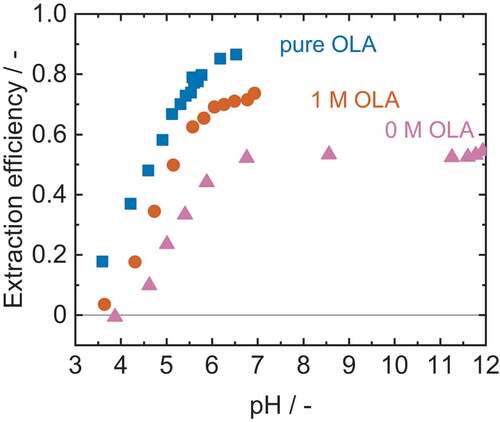
For solvent extraction of TDA with 1-octanol (0 M OLA), a maximum extraction efficiency of E = 54% was achieved. The partition coefficient determined in the experiments of logP = 0.07 only differs slightly from values of 0.03 reported in the literature.[Citation17] Reactive extraction with OLA, led to an increase in extraction efficiency with a maximum value of E = 86% when using pure OLA as organic phase. This increase of extraction efficiency is due to the already explained complex formation, making, reactive extraction with OLA generally applicable for the recovery of TDA.
When compared to the reactive extraction of TDA with D2EHPA,[Citation10] the use of OLA led to lower maximum extraction yields and seems inferior to the already proposed solution with D2EHPA. However, since D2EHPA has been proven to be non-biocompatible and requires a pH in the acidic range, the use of biocompatible OLA at pH 7 is a suitable alternative. In previous studies, it has already been demonstrated that small traces of 0.5 mM TDA already lead to a 20% reduction in the growth rate of P. putida KT2440.[Citation30] In the experiment shown above, by using pure OLA, the initial concentration of 2.4 mM could be reduced to 0.3 mM, but only since a low initial amine concentration was chosen. For higher amine concentrations, multistage operation should be envisaged to achieve higher extraction yields.
Conclusion
We have shown that, depending on the amine and pH value, solvent extraction or reactive extraction are appropriate separation methods to separate MDA and IPDA from an aqueous medium. MDA can be extracted to more than 97% by one-step extraction at neutral pH, thus making it compatible with potential up- and downstream bioprocessing. For IPDA, similar extraction efficiencies could be achieved with solvent extraction, but only with a pH > 12. The developed one-step IPDA reactive extraction enabled extraction efficiency higher than 98% at neutral pH. Expected other components of PU hydrolysates such as AA, sulfates, and phosphates only slightly affect the extraction efficiency by a minor pH shift of the extraction curve and allow selective amine extraction. Other possible PU monomers such as EG and BDO are co-extracted with a low extraction efficiency of E = 4% and E = 12%, respectively. IPDA extraction efficiencies show a pH shift to higher values in the presence of these components.
Furthermore, we were able to show with toxicity experiments that the remaining small amounts of MDA or IPDAwill not have a negative effect on the growth of P. putida and therefore do not cause any problems for the subsequent bio-upcycling. The developed extraction methods were additionally investigated for the extraction of TDA. An extraction efficiency of E = 52% was achieved for solvent extraction and an extraction efficiency of E = 86% for reactive extraction at neutral pH. In the work of work of Utomo et al.[Citation10] a higher extraction efficiency could be achieved, but the reactant D2EHPA used was not biocompatible compared to OLA in this work. Since it is already known that small traces of TDA lead to inhibition of fermentation of P. putida KT2440, there is a need for further research, by investigating a multi-step process or new solvents to further improve the TDA extraction efficiency.
In addition, the re-extraction of the amines from the organic phase should be investigated in a next step. A possibility for this is the transfer of the organic phase into an acidic phase, whereby the amine is re-extracted. This process is described in detail in the work of Doeker et al.[Citation21] Through the back extraction, valuable amines can be further purified from the aqueous phase by means of an evaporation. Moreover, a solvent recovery and reuse is possible.
LSEI_2193229_Supplementary_Material
Download PDF (394.8 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary data
Supplemental data for this article can be accessed online at https://doi.org/10.1080/07366299.2023.2193229.
Additional information
Funding
References
- Wolf, S.; Teitge, J.; Mielke, J.; Schütze, F.; Jaeger, C. The European Green Deal - More Than Climate Neutrality. Intereconomics. 2021, 56, 99–107. DOI: 10.1007/s10272-021-0963-z.
- Kemona, A.; Piotrowska, M. Polyurethane Recycling and Disposal: Methods and Prospects. Polymers. 2020, 12(8), 1752. DOI: 10.3390/polym12081752.
- Font, R.; Fullana, A.; Caballero, J.; Candela, J.; Garcı́a, A. Pyrolysis Study of Polyurethane. J. Anal. Appl. Pyrolysis. 2001, 58-59, 63–77. DOI: 10.1016/S0165-2370(00)00138-8.
- Zia, K. M.; Bhatti, H. N.; Ahmad Bhatti, I. Methods for Polyurethane and Polyurethane Composites, Recycling and Recovery: A Review. React. Funct. Polym. 2007, 67, 675–692. DOI: 10.1016/j.reactfunctpolym.2007.05.004.
- Yang, W.; Dong, Q.; Liu, S.; Xie, H.; Liu, L.; Li, J. Recycling and Disposal Methods for Polyurethane Foam Wastes. Proc.Environ. Sci. 2012, 16, 167–175. DOI: 10.1016/j.proenv.2012.10.023.
- Anuar Sharuddin, S. D.; Abnisa, F.; Wan Daud, W. M. A.; Aroua, M. K. A Review on Pyrolysis of Plastic Wastes. Energy Convers. Manage. 2016, 115, 308–326. DOI: 10.1016/j.enconman.2016.02.037.
- Datta, J.; Kopczyńska, P.; Simón, D.; Rodríguez, J. F. Thermo-Chemical Decomposition Study of Polyurethane Elastomer Through Glycerolysis Route with Using Crude and Refined Glycerine as a Transesterification Agent. J Polym. Environ. 2018, 26, 166–174. DOI: 10.1007/s10924-016-0932-y.
- Ellis, L. D.; Rorrer, N. A.; Sullivan, K. P.; Otto, M.; McGeehan, J. E.; Román-Leshkov, Y.; Wierckx, N.; Beckham, G. T. Chemical and Biological Catalysis for Plastics Recycling and Upcycling. Nat. Catal. 2021, 4, 539–556. DOI: 10.1038/s41929-021-00648-4.
- Wei, R.; Zimmermann, W. Microbial Enzymes for the Recycling of Recalcitrant Petroleum-Based Plastics: How Far are We? Microb. Biotechnol. 2017, 10, 1308–1322. DOI: 10.1111/1751-7915.12710.
- Utomo, R. N. C.; Li, W. J.; Tiso, T.; Eberlein, C.; Doeker, M.; Heipieper, H. J.; Jupke, A.; Wierckx, N.; Blank, L. M. Defined Microbial Mixed Culture for Utilization of Polyurethane Monomers. ACS Sustainable Chem. Eng. 2020, 8, 17466–17474. DOI: 10.1021/acssuschemeng.0c06019.
- Ballerstedt, H.; Tiso, T.; Wierckx, N.; Wei, R.; Averous, L.; Bornscheuer, U.; O’Connor, K.; Floehr, T.; Jupke, A.; Klankermayer, J.; et al. MIXed Plastics Biodegradation and UPcycling Using Microbial Communities: EU Horizon 2020 Project MIX-UP Started January 2020. Environ. Sci. Eur. 2021, 33, 99. DOI: 10.1186/s12302-021-00536-5.
- Shah, Z.; Gulzar, M.; Hasan, F.; Shah, A. A. Degradation of Polyester Polyurethane by an Indigenously Developed Consortium of Pseudomonas and Bacillus Species Isolated from Soil. Polym. Degrad. Stab. 2016, 134, 349–356. DOI: 10.1016/j.polymdegradstab.2016.11.003.
- Magnin, A.; Pollet, E.; Phalip, V.; Avérous, L. Evaluation of Biological Degradation of Polyurethanes. Biochem. Adv. 2020, 39, 107457. DOI: 10.1016/j.biotechadv.2019.107457.
- Sonnenschein, M. F. Polyurethanes: Science, Technology, Markets, and Trends; John Wiley & Sons: Hoboken, New Jersey, 2015.
- Wierckx, N. J. P.; Ballerstedt, H.; de Bont, J. A. M.; Wery, J. Engineering of Solvent-Tolerant Pseudomonas Putida S12 for Bioproduction of Phenol from Glucose. Appl. Environ. Microbiol. 2005, 71, 8221–8227. DOI: 10.1128/AEM.71.12.8221-8227.2005.
- Ackermann, Y. S.; Li, W. J.; de Hipt, L. O.; Niehoff, P. J.; Casey, W.; Polen, T.; Köbbing, S.; Ballerstedt, H.; Wynands, B.; O’Connor, K.; et al. Engineering Adipic Acid Metabolism in Pseudomonas Putida. Metab. Eng. 2021, 67, 29–40. DOI: 10.1016/j.ymben.2021.05.001.
- ECHA Database. European Chemicals Agency database. https://www.echa.europa.eu/.
- de Witt, J.; Ernst, P.; Gätgens, J.; Noack, S.; Hiller, D.; Wynands, B.; Wierckx, N. Characterization and Engineering of Branched Short-Chain Dicarboxylate Metabolism in Pseudomonas Reveals Resistance to Fungal 2-Hydroxyparaconate. Metab. Eng. 2023, 75, 205–216. DOI: 10.1016/j.ymben.2022.12.008.
- Bednarz, A.; Spieß, A. C.; Pfennig, A. Reactive and Physical Extraction of Bio-Based Diamines from Fermentation Media. J. Chem. Tech. Bio. 2017, 92, 1817–1824. DOI: 10.1002/jctb.5183.
- Rosinha Grundtvig, I. P.; Heintz, S.; Krühne, U.; Gernaey, K. V.; Adlercreutz, P.; Hayler, J. D.; Wells, A. S.; Woodley, J. M. Screening of Organic Solvents for Bioprocesses Using Aqueous-Organic Two-Phase Systems. Biochem. Adv. 2018, 36, 1801–1814. DOI: 10.1016/j.biotechadv.2018.05.007.
- Doeker, M.; Grabowski, L.; Rother, D.; Jupke, A. In situ Reactive Extraction with Oleic Acid for Process Intensification in Amine Transaminase Catalyzed Reactions. Green Chem. 2022, 24, 295–304. DOI: 10.1039/D1GC03289E.
- Akhtar, M. K.; Dandapani, H.; Thiel, K.; Jones, P. R. Microbial Production of 1-Octanol: A Naturally Excreted Biofuel with Diesel-Like Properties. Metab. Eng. Commun. 2015, 2, 1–5. DOI: 10.1016/j.meteno.2014.11.001.
- Doeker, M.; Hüttche, V.; Jupke, A. Reactive Extraction for the Recovery of Primary Amines from Aqueous Streams. Sep. Purif. Techn. 2021, 277, 118229. DOI: 10.1016/j.seppur.2020.118229.
- Kocks, C.; Krekel, C. M.; Gausmann, M.; Jupke, A. Determination of the Metastable Zone Width and Nucleation Parameters of Succinic Acid for Electrochemically Induced Crystallization. Crystals. 2021, 11, 1090. DOI: 10.3390/cryst11091090.
- Health Canada. Screening Assessment for Methylenediphenyl Diisocyanates and Methylenediphenyl Diamines: Chemical Abstracts Service Registry Numbers 101-68-8; 2536-05-2, 5873-54-1; 9016-87-9; 26447-40-5; 101-77-9; 25214-70-4 / Environment and Climate Change Canada, Health Canada. 2017.
- Tiso, T.; Ihling, N.; Kubicki, S.; Biselli, A.; Schonhoff, A.; Bator, I.; Thies, S.; Karmainski, T.; Kruth, S.; Willenbrink, A. L.; et al. Integration of Genetic and Process Engineering for Optimized Rhamnolipid Production Using Pseudomonas Putida. Front. Bioeng. Biotechnol. 2020, 8, 976. DOI: 10.3389/fbioe.2020.00976.
- Hansch, C.; Leo, A.; Hoekman, D. Hydrophobic, Electronic, and Steric Constants; American Chemical Society: Washington, DC, 1995.
- Qin, W.; Li, Z.; Dai, Y. Extraction of Monocarboxylic Acids with Trioctylamine: Equilibria and Correlation of Apparent Reactive Equilibrium Constant. Ind. Eng. Chem. Res. 2003, 42, 6196–6204. DOI: 10.1021/ie021049b.
- Rowe, R.; Sheskey, P. Handbook of Pharmaceutical Excipients 6th ed.; Quinn, M. Ed.; APhA (PhP) Pharmaceutical Press: London, 2009.
- Espinosa, M. J. C.; Blanco, A. C.; Schmidgall, T.; Atanasoff-Kardjalieff, A. K.; Kappelmeyer, U.; Tischler, D.; Pieper, D. H.; Heipieper, H. J.; Eberlein, C. Toward Biorecycling: Isolation of a Soil Bacterium That Grows on a Polyurethane Oligomer and Monomer. Front. Microbiol. 2020, 11, 404. DOI: 10.3389/fmicb.2020.00404.
