Abstract
Pre-mRNA splicing of RNA polymerase II transcripts is executed in the cell nucleus within a huge (21 MDa) and highly dynamic RNP machine – the supraspliceosome. Supraspliceosomes harbor all five spliceosomal U snRNPs, as well as protein splicing factors, and they are functional in splicing. Supraspliceosomes also harbor other components of pre-mRNA processing, such as the RNA-editing enzymes ADAR1 and ADAR2, cap-binding proteins, and 3’-end processing components. The supraspliceosome is composed of four native spliceosomes, each resembling the spliceosome assembled in vitro, and they are connected by the pre-mRNA (Sperling et al., 2008). The structure of the native spliceosome, at a resolution of 20 Å, was determined by cryo-EM (Azubel et al., 2004). A unique spatial arrangement of the spliceosomal U snRNPs within the native spliceosome emerged from in silico studies (Frankenstein et al., 2012). The model localizes the five U snRNPs mostly within the large subunit of the native spliceosome, requiring only minor conformation changes, and the components of the active core of the spliceosome are found sheltered deep within the large spliceosomal cavity. We use gold-tagged probes and cryo-EM to experimentally confirm this model. The supraspliceosome structure likely provides a platform for regulation of RNA-processing activities (e.g. alternative splicing). Indeed, all the phosphorylated splicing regulators SR proteins are predominantly found associated with supraspliceosomes. Furthermore, both the constitutively and alternatively spliced mRNAs of tested endogenous human Pol II transcripts are predominantly found in supraspliceosomes, and changes in alternative splicing occur within them (Sebbag-Sznajder et al., 2012). These findings substantiate the role of the supraspliceosome as the nuclear pre-mRNA processing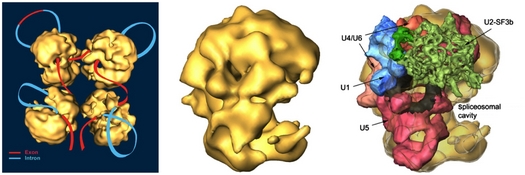 machine.
machine.
This research has been supported by the US NIH, grant GM079549 to R.S. and J.S., and the Helen and Milton Kimmelman Center for Biomolecular Structure and Assembly at the Weizmann Institute to J.S.
References
- Azubel , M. , Wolf , S. G. , Sperling , J. and Sperling , R. 2004 . Three-dimensional structure of the native spliceosome by cryo-electron microscopy . Molecular Cell , 15 : 833 – 839 .
- Frankenstein , Z. , Sperling , J. , Sperling , R. and Eisenstein , M. 2012 . A unique spatial arrangement of the snRNPs within the native spliceosome emerges from in silico studies . Structure , 20 : 1097 – 1106 .
- Sebbag-Sznajder , N. , Raitskin , O. , Angenitzki , M. , Sato , T. A. , Sperling , J. and Sperling , R. 2012 . Regulation of alternative splicing within the supraspliceosome . Journal of Structural Biology , 177 : 152 – 159 .
- Sperling , J. , Azubel , M. and Sperling , R. 2008 . Structure and function of the pre-mRNA splicing machine . Structure , 16 : 1605 – 1615 .