Abstract
Background
Though the previous genome-wide association studies found the association between HLA alleles and rosacea in the European populations, the data is lacking among the Asians. Moreover, neutrophils are important in the immune-related mechanism of rosacea, and dyslipidemia is closely related to rosacea. We aimed to explore the association between HLA genes and rosacea in Chinese rosacea patients, as well as the mediation effect of neutrophils, high-density lipoprotein (HDL) and low-density lipoprotein (LDL) on the relationship between HLA genes and rosacea.
Methods
A total of 249 rosacea and 150 controls were ranked by the international investigator global rosacea severity scores. HLA genes, neutrophils, HDL, and LDL were detected. And their mediation effects on the relationship between HLA and rosacea risk or severity were analysed.
Results
HLA-DQB1*03:03 allele (OR = 41.89, 95% CI: 9.80 ∼ 179.09, p = 4.7*10−7), HLA-DQB1*04:02 allele (OR = 0.16, 95% CI: 0.03 ∼ 0.81, p = 0.026) and HLA-DQB1*03:03/05:02 genotype (OR = 5.57, 95% CI: 1.13 ∼ 27.52, p = 0.0351) were significantly associated with rosacea. Moreover, HLA-DQB1*03:03 allele (b = 1.434, SE = 0.217, p = 2.0*10−10), HLA-DQB1*05:01 allele (b = 0.894, SE = 0.33520, p = 0.008) and HLA-DQB1*03:03/06:01 genotype (b = 0.998, SE = 0.472, p = 0.040) were positively associated with rosacea severity. Furthermore, we found both neutrophils and HDL, instead of LDL, have mediation effects on the relationship between HLA-DQB1*03:03 and risk or severity of rosacea.
Conclusions
We discovered novel susceptible HLA alleles for rosacea in the Chinese population, and disclosed the mediation effect of neutrophils and HDL on the relationship between HLA-DQB1 and rosacea, implying a possible correlation between rosacea and inflammatory or metabolic factors, providing hints for future studies in the mechanism of rosacea.
HLA-DQB1*03:03 allele, HLA-DQB1*04:02 allele and HLA-DQB1*03:03/05:02 genotype were significantly associated with rosacea.
HLA-DQB1*03:03 allele, HLA-DQB1*05:01 allele and HLA-DQB1*03:03/06:01 genotype were positively associated with rosacea severity.
Neutrophils and HDL have mediation effects on the relationship between HLA-DQB1*03:03 and risk or severity of rosacea.
Key messages
Introduction
Rosacea is a chronic inflammatory disease that primarily affects facial skin, which has a negative effect on patients’ quality of life and mental health [Citation1]. The major features of rosacea include erythema, telangiectasia, phymatous changes, ocular manifestations, and irritative sensations [Citation2–4]. The prevalence of rosacea in China is approximately 3.48% based on a cross-section study [Citation5].
More than one-third of rosacea patients have family history inclination [Citation6–9]. Transcriptome profile analysis discovered that different subtypes of rosacea have certain genetic profiles [Citation10,Citation11]. Genome-wide association studies (GWAS) revealed some human leukocyte antigen (HLA) alleles were associated with the risk of rosacea among the Europeans. For example, HLA-DMA/B was correlated with immuno-inflammation phenotypes in rosacea, which was consistent with the inflammatory pathogenesis of rosacea [Citation12,Citation13]. However, the genetic background of rosacea has been lacking in Asian populations.
Besides, immune dysfunction is one of the dominating mechanisms of rosacea. Neutrophils, making up the majority of innate immune effector cells, react immediately against acute inflammation and immune reactions when exposed to pathogens [Citation14]. Vast research confirmed that rosacea lesions are infiltrated with abundant neutrophils, suggesting a vital role of neutrophils played in the immune-related mechanism of rosacea. HLA is essential to immune reactions by presenting antigens and recruiting immunocytes including neutrophils. Besides, HLA, which presents antigens from extracellular sources, may explain the connection between microbes and rosacea and could participate in immune reactions of rosacea [Citation13]. Hence, we speculate that HLA might mediate immune dysfunction in rosacea through mediating the expression of inflammatory cells, especially neutrophils. Therefore, this study aimed to investigate the mediation effect of neutrophils on the association between HLA and rosacea, if any.
Interestingly, rosacea is closely related to metabolic factors such as hypertension and dyslipidemia [Citation15–17]. As reported, blood lipids, including total cholesterol, low-density lipoprotein and triglyceride, in rosacea patients were much higher than in healthy controls and obese patients were more susceptible to rosacea [Citation17–20]. High-density lipoprotein (HDL) and low-density lipoprotein (LDL) are the main cholesterol-carrying lipoproteins [Citation21]. Low level of HDL increases the risk of cardiovascular disease (CVD), since HDL has anti-inflammatory effects [Citation22]. In contrast, down-regulation of LDL by medicines prevents atherosclerotic inflammation and lowers the incidence of CVD [Citation23–27]. Interestingly, HLA alleles relate to HDL or LDL in patients with cardiovascular disease, atherogenesis, and rheumatoid arthritis. For instance, HLA-DR expression regulates LDL expression and immune-inflammatory cell content. Besides, genetic variants of HLA-DQB1 connected with human longevity were associated with LDL/HDL ratio in a long-lived population [Citation21,Citation28–31]. Accordingly, we wonder if HLA alleles affect rosacea through mediating the expression of HDL and LDL.
In this study, we aimed at exploring the HLA typing among Chinese rosacea patients and investigating the mediation effect of inflammatory (neutrophils in peripheral blood) and metabolic (HDL, LDL) factors on the association between the susceptible HLA alleles and rosacea.
Materials and methods
Study design and population
This study was conducted from November 2017 to November 2019, in Xiangya Hospital of Central South University, Changsha. A total of 249 rosacea patients were recruited from the department of dermatology, diagnosed and divided into phenotypes of erythematotelangiectatic (ETR), papulopustular (PPR), and phymatous rosacea (PhR) by the 2004 diagnostic criteria determined by the National Rosacea Society Expert Committee [Citation32]. 100 healthy controls, without a history of medication within the past three months, dieting, pregnancy, lactation, rosacea or other diseases, were recruited from the physical examination center. Patients with a history of medication within the past three months, dieting, pregnancy, or lactation, systemic diseases, specifically, cardiovascular disease, severe infections, mental illness, or other skin diseases (e.g. psoriasis and atopic dermatitis) that may interfere with the assessment of rosacea were excluded. The disease severity of rosacea was determined by the international investigator global rosacea severity scores (IGA scores) [Citation33]. All assessments were completed by two dermatologists independently. Laboratory tests were conducted in Xiangya Hospital, including white blood cell count analysed by flow cytometer and serum HDL/LDL detected by colorimetry.
All participants signed written informed consents. This research was approved by the ethics review board of Xiangya Hospital Central South University and the approval number is NO.201611608, which covers the Declaration of Helsinki requirements.
DNA extraction and quality control
Peripheral venous blood was drawn from participants after 12 h overnight fasting. Centrifuging blood samples at 4000 g for 30 min to separate haemocytes and plasma. Genomic DNA was extracted from human blood cell samples by HiPure Blood & Tissue DNA Kit (Magen, Cat#D3018-03). 1% of agarose gels were applied to test the degradation of DNA. The quality control of DNA was monitored by the NanoDrop spectrophotometer (ND-2000, Thermo Fisher Scientific).
HLA genotyping and imputation
Alleles from five HLA genes (HLA-A, HLA-B, HLA-C, HLA-DQB1, HLA-DRB1) were genotyped based on polymerase chain reaction with sequence-based typing (PCR-SBT) by TBG HLAssure SE DQB1 Locus SBT Kit (TBG Biotechnology Xiamen Inc). HLA allele results were analysed by supporting software AccuType (BioSoft, Oklahoma, USA).
Statistical analysis
The distribution of gender between patients and controls was compared by Chi-squared test, and other base characteristics by independent-sample t-test. When analyzing the relationship between the alleles or genotypes and rosacea, we took the highest frequent alleles or genotypes as the reference and calculated the odds ratios (OR) and 95% confidence intervals (CI) through logistic regression. The association between the alleles or genotypes and rosacea disease severity (IGA score) was analysed by linear regression. When the total associations (c) between the allele/genotype (independent X) and rosacea, rosacea severity was significant, the mediation analysis would be considered. The potential mediators (M) were neutrophils, HDL and LDL. Then the association (a) between X and each of the mediators was estimated, as well as the association between each of the mediators and Y after controlling for X (b) and the direct association (c′) between X and Y after controlling for M. When a and b were both significant associations, the indirect association (a × b) was calculated as the mediation effect. The percentage of the mediation effects equals to (a × b)/(a × b + c′). The pathways were all tested using bias-corrected bootstrapped 95% confidence intervals (b = 5000) and would be considered significant when a bootstrapped confidence interval does not include zero. All analyses were performed using SPSS and were considered significant when p < 0.05.
Results
HLA allele, genotype and risk of rosacea
Alleles of five HLA genes (HLA-A, HLA-B, HLA-C, HLA- DRB, HLA-DQB1) were genotyped among 100 rosacea patients (31.90 ± 10.53 years old) and 100 health controls (35.27 ± 8.85 years old), among which HLA-A*11:01 (7.4% in rosacea, 1.6% in controls), HLA-B*40:01 (8% in rosacea, 11.5% in controls), HLA-C*01:02 (12% in rosacea, 10% in controls), HLA-DRB*09:01 (10% in rosacea, 8.3% in controls), HLA-DQB1*03:01 (7.8% in rosacea, 11.3% in controls) acquired the highest frequency in each gene. HLA-A, HLA-B and HLA- DRB were not relevant to rosacea. HLA-C*03:04 allele was negatively associated with risk of rosacea (4.8% in rosacea, and 8.5% in controls; OR = 0.47, CI: 0.23 ∼ 0.94, p = 0.033). Contrarily, HLA-DQB1*03:03 (10.8% in rosacea, and 8.5% in controls; OR = 1.84, CI: 0.97 ∼ 3.49, p = 0.063) and HLA-DQB1*06:02 (3.5% in rosacea, and 2.3% in controls; OR = 2.26, CI: 0.87 ∼ 5.86, p = 0.094) alleles were positively associated with rosacea with marginal significance (Supplementary Table 1). Besides, HLA-DQB1 03:03/03:03 (3.0% in cases, and 0.5% in controls; OR = 12.00, 95%CI: 1.12 ∼ 128.84, p = 0.040) genotypes were positively associated with risk of rosacea, and HLA-DQB1 03:03/05:02 (3.5% in cases, and 1.5% in controls; OR = 4.67, 95%CI: 0.83 ∼ 26.24, p = 0.080) was found on the brink of significance correlated with rosacea (Supplementary Table 2). However, genotypes of other HLA genes had no significant associations with the risk of rosacea. Hence, HLA-DQB1 may be a potential risk gene of rosacea.
HLA-DQB1 allele, genotype and risk, severity (IGA) of rosacea
To further explore the association between HLA-DQB1 alleles and rosacea, we enlarged the sample size to 399 individuals, including 249 rosacea patients and 150 healthy controls. The demographic information of selected subjects was listed in and there was no significant difference between case and control groups in the distribution of gender or age. The proportion of ETR, PPR, and PhR rosacea patients were 58.2%, 35.7%, and 6.0%, respectively ().
Table 1. The demographic and clinical features of the rosacea cases and controls.
Twelve alleles and 9 genotypes were detected on HLA-DQB1 among 399 individuals, of which the frequency of HLA-DQB1*03:01 was the highest (20.4% in controls). The HLA-DQB1*03:03 allele was positively associated with risk of rosacea (12.8% in cases, and 7.0% in controls; OR = 41.89, 95% CI: 9.80 ∼ 179.09, p = 4.7*10−7), while HLA-DQB1*04:02 was negatively associated with risk of rosacea (0.3% in cases, and 0.9% in controls; OR = 0.16, 95% CI: 0.03 ∼ 0.81, p = 0.026). Besides, the HLA-DQB1 genotype 03:03/05:02 was positively associated with risk of rosacea (3.3% in cases, and 0.8% in controls; OR = 5.57, 95% CI: 1.13 ∼ 27.52, p = 0.0351) after enlarging sample size. Other alleles or genotypes had no significant associations with the risk of rosacea ().
Table 2. Association between HLA-DQB1 allele, genotype and rosacea *.
Except for risk of rosacea, alleles and genotypes of HLA-DQB1 were strongly associated with disease severity of rosacea (). HLA-DQB1*03:03 and HLA-DQB1*05:01 were positively correlated with disease severity (b = 1.434, p = 2.0*10−10; b = 0.894, p = 0.008, respectively). In addition, HLA-DQB1*03:03/06:01 genotype had a positive correlation with IGA scores (b = 0.998, p = 0.040).
Table 3. Association between HLA-DQB1 allele, genotype and rosacea severity.
Mediation role of neutrophils, HDL and LDL on the relationship between HLA-DQB1 allele/genotype and rosacea risk or rosacea severity
Since HLA-DQB1 might be a potential risk gene, and HLA locus could regulate neutrophils, HDL and LDL, we herein collected the laboratory tests of all participants to explore the association between HLA-DQB1 allele/genotype, neutrophils, HDL, LDL, rosacea risk, and rosacea severity (, , Supplementary Table 3).
Figure 1. Mediation analysis for HLA-DQB1*03:03, neutrophils, HDL and rosacea risk or severity. HDL: high-density lipoprotein; IGA: disease severity of rosacea.
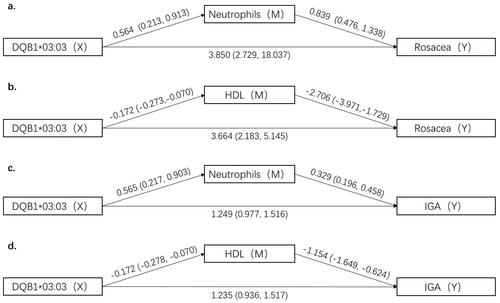
Table 4. Association between HLA-DQB1 gene, neutrophils, HDL, LDL, rosacea and rosacea severity (IGA).
In the associations between HLA-DQB1 and the potential mediators for rosacea risk, HLA-DQB1*03:03 was positively associated with neutrophils (β = 0.564, 95% CI: 0.213 ∼ 0.913), while negatively correlated with HDL (β= −0.172, 95% CI: −0.273∼−0.070). As for rosacea severity, HLA-DQB1*03:03 was positively associated with neutrophils (β = 0.565, 95% CI: 0.217 ∼ 0.903), while negatively correlated with HDL (β= −0.172, 95% CI: −0.278∼−0.070). As for the association between potential mediators and rosacea risk, or severity, neutrophils were positively associated with rosacea risk (β = 0.839, 95% CI: 0.476 ∼ 1.338) and disease severity (β = 0.329, 95% CI: 0.196 ∼ 0.458), while HDL was negatively associated with rosacea (β= −2.706, 95% CI: −3.971∼−1.729) and IGA score (β= −1.154, 95% CI: −1.649∼−0.624).
Furthermore, we analysed the mediation effects of neutrophils and HDL on the relationship between the HLA-DQB1 allele and rosacea risk, or severity (). The mediation effect of neutrophils on the relationship between HLA-DQB1*03:03 and rosacea risk was 0.47 (95% CI: 0.16 ∼ 0.92), and the mediation ratio was 10.9%. The mediation effect of neutrophils on the relationship between HLA-DQB1*03:03 and rosacea severity was 0.18 (95% CI: 0.06 ∼ 0.34, mediation ratio = 12.59%). For HDL, the mediation effect was 0.46 (95% CI: 0.18 ∼ 0.86) on the relationship of HLA-DQB1*03:03 and rosacea risk (mediation ratio = 11.17%), 0.20 (95% CI: 0.07 ∼ 0.37) on the relationship of HLA-DQB1*03:03 and rosacea severity (mediation ratio = 13.89%).
Table 5. Mediation of neutrophils and HDL, LDL on the relationship between HLA-DQB1 allele and rosacea or rosacea severity.
Discussion
In this study, we identified new susceptible HLA alleles of rosacea, especially HLA-DQB1*03:03, among Chinese population, and clarified the mediation effect of neutrophils and HDL in the association between HLA-DQB1*03:03 and rosacea.
Genetic background was considered important in the incidence and severity of rosacea by twin studies [Citation10]. GWAS in Europeans revealed an association between HLA alleles and rosacea risk/severity, including HLA-DRB1*03:01, HLA-DQB1*02:01, and HLA-DQA1*05:01 [13]. However, relevant research in Asia has been absent. We identified HLA-C*03:04 and HLA-DQB1*04:02 as protective alleles, while HLA-DQB1*03:03 allele and HLA-DQB1*03:03/05:02 genotype as risk factors. Besides, HLA-DQB1*03:03, HLA-DQB1*05:01, and HLA-DQB1*03:03/06:01 were positively associated with rosacea severity. The discrepancy between our study and the previous studies may attribute to the differences in ethnicity, or the relatively limited sample size. Interestingly, HLA-DQB1*03:03, which was associated with both risk and severity of rosacea in our study, was reported to have linkage with various immune diseases, including pemphigus vulgaris, pemphigus foliaceous, multiple sclerosis and auto-immune thyroid disease [Citation34–36]. Hence, we speculated that HLA-DQB1*03:03 was involved in the mechanism of rosacea through inducing immunologic disorder. In brief, our study further confirmed the concept of an inflammatory genetic component in rosacea.
Lesions of rosacea are infiltrated by inflammatory cells, especially neutrophils, which was confirmed by immunohistochemistry and transcriptome analysis [Citation37,Citation38]. Moreover, topical medications like metronidazole alleviate disease condition by inhibiting neutrophils [Citation39–41]. However, obtaining neutrophils from the lesions is invasive and the fluctuation of neutrophil levels in peripheral blood reflects the changes in the individual’s overall inflammatory status to some extent. Hence, we explored the mediation effect of neutrophils in peripheral blood on the relationship between HLA-DQB1*03:03 and rosacea risk or severity, revealing the mediation effect at 10.9% and 12.59%, respectively. This indicates that neutrophils play a role in the HLA-related immune mechanism in rosacea. Since neutrophils can be stimulated and activated by MHC class II alleles [Citation42–46], we presume that HLA-DQB1*03:03 might recruit and stimulate neutrophils in rosacea. Still, the underlying mechanism needs further exploration.
Rosacea has been reported to be highly related to metabolic factors, such as dyslipidemia [Citation15–17], indicating that metabolic pathway might get involved in rosacea. HLA-DQB1 alleles, proved to be associated with risk and severity of rosacea in our study, have been found related to abnormal HDL-c levels in a cross-sectional study among the Chinese long-living population [Citation21]. Consequently, we explored the mediating effects of HDL and LDL in HLA-DQB1-influenced risk and severity of rosacea. HDL expression was found negatively correlated with HLA-DQB1*03:03 and rosacea risk or severity, indicating a protective role of HDL in rosacea, with the mediation effect on the relationship between HLA-DQB1*03:03 and rosacea risk/severity as 11.17% and 13.89%, respectively. The association between rosacea and abnormal lipid metabolism provides a novel hint for the mechanism of rosacea. HDL levels are negatively associated with gene expression of human cathelicidin (LL-37), a key peptide involved in the pathogenesis of rosacea, which explained the possible relevance between HDL and rosacea [Citation19,Citation47]. In addition, human serum paraoxonase (PON1) with low activity enables HDL to be more susceptible to oxidation, which may increase the risk of rosacea [Citation48–50]. Conclusively, HLA-DQB1 alleles might induce rosacea through inhibiting HDL expression, which needs further exploration.
The strengths of our study were the HLA typing in Asian rosacea populations and the mediation analysis of both inflammatory and metabolic factors, which fulfil the blank of the genetic research of Asian groups and provide new hints for the mechanism of rosacea. The limitation of our research is the insufficient sample sizes. Though we recruited nearly four hundred people, the positive number of each HLA allele was still limited.
In conclusion, novel HLA alleles are associated with rosacea risk and severity in the Chinese group. HLA-DQB1*03:03 is an important genetic predisposition for rosacea. Besides, we explored the mediation effect of both neutrophils and HDL on the relationship between HLA-DQB1 alleles and rosacea risk or severity, implicating that inflammatory and metabolic-related factors might get involved in the mechanism of rosacea under the genetic basis. This discovery provides new directions for targeted therapy and comprehension of rosacea.
Author contributions
Conception and design: J.L. and Y.T. Financial support: J.L. and H.X. Collection and assembly of data: W.X, Q.Z., T.L., and Z.D. Data analysis and interpretation: X.H. and Y.T. Manuscript writing and revision: W.X. Final approval of manuscript: J.L. and Y.T.
Supplemental Material
Download MS Word (20.5 KB)Acknowledgments
The authors thank all coordinators, dermatologists, and investigators that participated in the field survey. Specific consent for publication of all participants was obtained.
Disclosure statement
The authors declare no conflicts of interests.
Data availability statement
Data are contained within this article or its supplementary material.
Additional information
Funding
References
- van Zuuren EJ. Rosacea. N Engl J Med. 2017;377(18):1530–1764.
- Deng Y, Peng Q, Yang S, et al. The rosacea-specific quality-of-Life instrument (RosQol): revision and validation among Chinese patients. PLoS One. 2018;13(2):e0192487.
- Gallo RL, Granstein RD, Kang S, et al. Standard classification and pathophysiology of rosacea: the 2017 update by the national rosacea society expert committee. J Am Acad Dermatol. 2018;78(1):148–155.
- Thiboutot D, Anderson R, Cook-Bolden F, et al. Standard management options for rosacea: the 2019 update by the national rosacea society expert committee. J Am Acad Dermatol. 2020;82(6):1501–1510.
- Li J, Wang B, Deng Y, et al. Epidemiological features of rosacea in Changsha, China: a population-based, cross-sectional study. J Dermatol. 2020;47(5):497–502.
- Two AM, Wu W, Gallo RL, et al. Rosacea: part I. Introduction, categorization, histology, pathogenesis, and risk factors. J Am Acad Dermatol. 2015;72(5):749–758. quiz 59–60.
- Abram K, Silm H, Maaroos HI, et al. Risk factors associated with rosacea. J Eur Acad Dermatol Venereol. 2010;24(5):565–571.
- Steinhoff M, Schauber J, Leyden JJ. New insights into rosacea pathophysiology: a review of recent findings. J Am Acad Dermatol. 2013;69(6 Suppl 1):S15–S26.
- Elsaie ML, Choudhary S. Updates on the pathophysiology and management of acne rosacea. Postgrad Med. 2009;121(5):178–186.
- Aldrich N, Gerstenblith M, Fu P, et al. Genetic vs environmental factors that correlate with rosacea: a cohort-based survey of twins. JAMA Dermatol. 2015;151(11):1213–1219.
- Awosika O, Oussedik E. Genetic predisposition to rosacea. Dermatol Clin. 2018;36(2):87–92.
- Aponte JL, Chiano MN, Yerges-Armstrong LM, et al. Assessment of rosacea symptom severity by genome-wide association study and expression analysis highlights immuno-inflammatory and skin pigmentation genes. Hum Mol Genet. 2018;27(15):2762–2772.
- Chang ALS, Raber I, Xu J, et al. Assessment of the genetic basis of rosacea by genome-wide association study. J Invest Dermatol. 2015;135(6):1548–1555.
- Mayadas T, Cullere X, Lowell C. The multifaceted functions of neutrophils. Annu Rev Pathol. 2014;9:181–218.
- Chen Q, Shi X, Tang Y, et al. Association between rosacea and cardiometabolic disease: a systematic review and meta-analysis. J Am Acad Dermatol. 2020;83(5):1331–1340.
- Hua T, Chung P, Chen Y, et al. Cardiovascular comorbidities in patients with rosacea: a nationwide case-control study from Taiwan. J Am Acad Dermatol. 2015;73(2):249–254.
- Duman N, Ersoy Evans S, Atakan N. Rosacea and cardiovascular risk factors: a case control study. J Eur Acad Dermatol Venereol. 2014;28(9):1165–1169.
- Akin Belli A, Kara A, Ozbas Gok S. Can hematologic parameters be an indicator of metabolic disorders accompanying rosacea? Acta Dermatovenerol Croat. 2017;25(2):145–150.
- Akin Belli A, Ozbas Gok S, Akbaba G, et al. The relationship between rosacea and insulin resistance and metabolic syndrome. Eur J Dermatol. 2016;26(3):260–264.
- Hirt P, Castillo D, Yosipovitch G, et al. Skin changes in the obese patient. J Am Acad Dermatol. 2019;81(5):1037–1057.
- Yang F, Sun L, Zhu X, et al. Identification of new genetic variants of HLA-DQB1 associated with human longevity and lipid homeostasis-a cross-sectional study in a Chinese population. Aging (Albany NY). 2017;9(11):2316–2333.
- Rosenson R, Brewer H, Ansell B, et al. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat Rev Cardiol. 2016;13(1):48–60.
- Lewington S, Whitlock G, Clarke R, et al. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet. 2007;370(9602):1829–1839.
- Ridker P. LDL cholesterol: controversies and future therapeutic directions. Lancet. 2014;384(9943):607–617.
- Baigent C, Blackwell L, Emberson J, et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376:1670–1681.
- Mihaylova B, Emberson J, Blackwell L, et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 2012;380:581–590.
- Tawakol A, Fayad Z, Mogg R, et al. Intensification of statin therapy results in a rapid reduction in atherosclerotic inflammation: results of a multicenter fluorodeoxyglucose-positron emission tomography/computed tomography feasibility study. J Am Coll Cardiol. 2013;62(10):909–917.
- Toms T, Panoulas V, Smith J, et al. Rheumatoid arthritis susceptibility genes associate with lipid levels in patients with rheumatoid arthritis. Ann Rheum Dis. 2011;70(6):1025–1032.
- Davis L, Whitfield E, Cannon G, et al. Association of rheumatoid arthritis susceptibility gene with lipid profiles in patients with rheumatoid d arthritis. Rheumatology (Oxford). 2014;53(6):1014–1021.
- Bobryshev Y, Andreeva E, Mikhailova I, et al. Correlation between lipid deposition, immune-inflammatory cell content and MHC class II expression in diffuse intimal thickening of the human aorta. Atherosclerosis. 2011;219(1):171–183.
- Nikpay M, McPherson R. Convergence of biomarkers and risk factor trait loci of coronary artery disease at 3p21.31 and HLA region. NPJ Genom Med. 2021;6(1):12.
- Wilkin J, Dahl M, Detmar M, et al. Standard grading system for rosacea: report of the national rosacea society expert committee on the classification and staging of rosacea. J Am Acad Dermatol. 2004;50(6):907–912.
- Taieb A, Khemis A, Ruzicka T, Ivermectin Phase III Study Group, et al. Maintenance of remission following successful treatment of Papulopustular rosacea with Ivermectin 1% cream vs. metronidazole 0.75% cream: 36-week extension of the ATTRACT randomized study. J Eur Acad Dermatol Venereol. 2016;30(5):829–836.
- Zhang S, Zhou X, Zhou X, et al. Subtype-specific inherited predisposition to pemphigus in the Chinese population. Br J Dermatol. 2019;180(4):828–835.
- Čierny D, Lehotský J, Kantorová E, et al. The HLA-DRB1 and HLA-DQB1 alleles are associated with multiple sclerosis disability progression in Slovak population. Neurol Res. 2018;40(7):607–614.
- Ramgopal S, Rathika C, Padma Malini R, et al. Critical amino acid variations in HLA-DQB1* molecules confers susceptibility to autoimmune thyroid disease in South India. Genes Immun. 2019;20(1):32–38.
- Buddenkotte J, Steinhoff M. Recent advances in understanding and managing rosacea. F1000Res. 2018;7:1885. F1000 Faculty Rev-
- Buhl T, Sulk M, Nowak P, et al. Molecular and morphological characterization of inflammatory infiltrate in rosacea reveals activation of Th1/Th17 pathways. J Invest Dermatol. 2015;135(9):2198–2208.
- Bertino B, Blanchet-Réthoré S, Thibaut de Ménonville S, et al. Brimonidine displays anti-inflammatory properties in the skin through the modulation of the vascular barrier function. Exp Dermatol. 2018;27(12):1378–1387.
- Fernandez-Obregon A, Patton D. The role of Chlamydia pneumoniae in the etiology of acne rosacea: response to the use of oral azithromycin. Cutis. 2007;79:163–167.
- Miyachi Y, Imamura S, Niwa Y. Anti-oxidant action of metronidazole: a possible mechanism of action in rosacea. Br J Dermatol. 1986;114(2):231–234.
- Matsushima H, Geng S, Lu R, et al. Neutrophil differentiation into a unique hybrid population exhibiting dual phenotype and functionality of neutrophils and dendritic cells. Blood. 2013;121(10):1677–1689.
- Oehler L, Majdic O, Pickl W, et al. Neutrophil granulocyte-committed cells can be driven to acquire dendritic cell characteristics. J Exp Med. 1998;187(7):1019–1028.
- Vono M, Lin A, Norrby-Teglund A, et al. Neutrophils acquire the capacity for antigen presentation to memory CD4+ T cells in vitro and ex vivo. Blood. 2017;129(14):1991–2001.
- Radsak M, Iking-Konert C, Stegmaier S, et al. Polymorphonuclear neutrophils as accessory cells for T-cell activation: major histocompatibility complex class II restricted antigen-dependent induction of T-cell proliferation. Immunology. 2000;101(4):521–530.
- Iking-Konert C, Vogt S, Radsak M, et al. Polymorphonuclear neutrophils in Wegener's granulomatosis acquire characteristics of antigen presenting cells. Kidney Int. 2001;60(6):2247–2262.
- Benachour H, Zaiou M, Samara A, et al. Association of human cathelicidin (hCAP-18/LL-37) gene expression with cardiovascular disease risk factors. Nutr Metab Cardiovasc Dis. 2009;19(10):720–728.
- Ayub A, Mackness M, Arrol S, et al. Serum paraoxonase after myocardial infarction. ATVB. 1999;19(2):330–335.
- Shih D, Gu L, Xia Y, et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394(6690):284–287.
- Mackness M, Harty D, Bhatnagar D, et al. Serum paraoxonase activity in familial hypercholesterolaemia and insulin-dependent diabetes mellitus. Atherosclerosis. 1991;86(2-3):193–199.
