Abstract
Infectious diseases are a major threat for human and animal health worldwide. Artificial Intelligence (AI) combined algorithms including Machine Learning and Big Data analytics have emerged as a potential solution to analyse diverse datasets and face challenges posed by infectious diseases. In this commentary we explore the potential applications and limitations of ML to management of infectious disease. It explores challenges in key areas such as outbreak prediction, pathogen identification, drug discovery, and personalized medicine. We propose potential solutions to mitigate these hurdles and applications of ML to identify biomolecules for effective treatment and prevention of infectious diseases. In addition to use of ML for management of infectious diseases, potential applications are based on catastrophic evolution events for the identification of biomolecular targets to reduce risks for infectious diseases and vaccinomics for discovery and characterization of vaccine protective antigens using intelligent Big Data analytics techniques. These considerations set a foundation for developing effective strategies for managing infectious diseases in the future.
KEY MESSAGES
Infectious diseases are a major challenge worldwide
Artificial Intelligence (AI) combined algorithms have emerged as a potential solution to analyse diverse datasets and face challenges posed by infectious diseases
Future directions include applications of ML to identify biomolecules for effective treatment and prevention of infectious diseases
Introduction
The rise of ever-changing infectious diseases, like the recent coronavirus disease 2019 (COVID-19) pandemic, emphasises the urgent need for innovative tools to combat their spread and mitigate their impact. Machine learning (ML), which is part of artificial intelligence (AI) has emerged as a potential solution due to its ability to analyse diverse datasets [Citation1,Citation2].
Many studies in the literature have explored the applications of ML in managing infectious diseases. These applications range from predicting outbreaks and tracing transmission routes to aiding diagnosis and developing treatment and preventive strategies. For example, Park et al. [Citation3] developed a model that combined Deep Learning (DL) with ML models to enhance disease prediction accuracy based on laboratory tests. Their optimized ensemble model achieved a 92% prediction accuracy, showing precision and recall rates for diseases like acute hepatitis B, malaria, meningitis among other [Citation3]. Another study reported that ML showed promising results in predicting poor outcomes of bloodstream infections, with an area under the receiver-operating characteristics curve of 0.82 [Citation4], which supports the potential of ML models in early infection management.
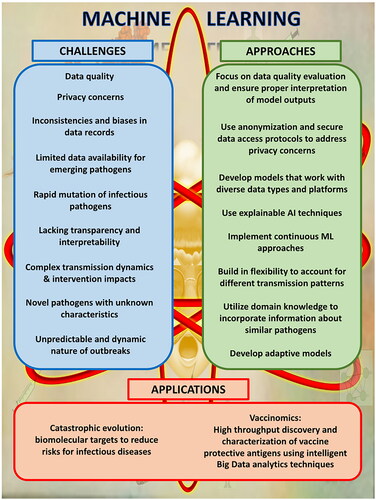
However, it is important to consider that the implementation of ML in infectious disease management comes with its set of challenges [Citation5]. These challenges include the requirement for high-quality data to ensure that the ML models can be applicable across different populations and pathogens. The ML models often face difficulties with overfitting and lacking interpretability. Biased data can result in skewed healthcare outcomes. Integrating these models into existing healthcare workflows poses additional obstacles. Given the evolution of agents and the variability in disease presentation, models can quickly become outdated, thus requiring continuous updates and validation [Citation5,Citation6].
The aim of this commentary is to explore the potential applications and barriers to ML, specifically in the context of infectious disease management. It explores challenges in key areas such as outbreak prediction, pathogen identification, drug discovery, and personalized medicine. Additionally, it proposes potential solutions to mitigate these hurdles and applications of ML. These considerations set a foundation for fair strategies in managing infectious diseases in the future.
From an epidemiological point of view, multiple research projects have been done focused on the prediction and prevention of epidemiological outbreaks using different and out of the box approaches. The ML algorithms have been found to be useful in evaluating outbreak risk of a zone, when diseases such as vector-borne diseases are influenced by environmental or meteorological factors. For example, in the case of malaria, researchers predicted the risk of outbreaks by using meteorological data such as rainfall, maximum and minimum temperatures, and other variables [Citation7]. Similar approaches have been applied combined with socio-economic variables for dengue [Citation8], and COVID-19 [Citation9] allowing authorities to act with preventing measures.
On the prevention line but with a different approach, multiple efforts using social media data have been used to manage transmissible diseases. Tracking the evolution of diseases can be done in real-time by taking advantage of social networks immediacy. Using tools as text mining, social media analysis (SMA) or sentiment analysis is possible to find work on early detection of outbreaks, detection of infected individuals or to predict transmission patterns or risks based on interaction between users [Citation10]. These works have achieved good accuracy by using text as input (mainly from Twitter) and evaluating the presence of some key words (such as the main synonyms). While there are concerns about privacy and usability, it could be a useful tool for precise detection. Also, there is another set of ML tools that have been trained to manage outbreaks that have started or even pandemics. For example, Moulaei et al. [Citation11] developed a tool to predict infected individuals with a higher risk of developing a serious health condition, opening the door to better hospital management in case of saturation. A summary of these studies with more details on the ML models is disclosed in .
Table 1. Summary of reviewed studies on the use of ML in infectious diseases.
Methodology
To ensure a comprehensive understanding of the application of ML in managing infectious diseases, we performed a multi-database search in PubMed (https://pubmed.ncbi.nlm.nih.gov), Scopus (https://www.scopus.com/), and Web of Science (https://clarivate.com/products/scientific-and-academic-research/research-discovery-and-workflow-solutions/webofscience-platform/), complemented by conference proceedings and grey literature to capture a wide range of discussions on ML applications in infectious diseases. The search strategy was built around a combination of keywords related to "machine learning", "infectious diseases", "predictive modelling", "data challenges", and specific terms related to the diseases discussed such as "SARS-CoV-2", "Ebola", and "influenza". Boolean operators (AND, OR) were used to refine the search results and ensure relevance. We established clear inclusion and exclusion criteria to focus on articles that directly address ML applications, challenges, and opportunities within infectious disease management. Data extraction was followed by a critical synthesis of the articles, focusing on identifying common themes, technological gaps, and the integration of diverse scientific perspectives.
Barriers to ML in infectious diseases
The application of ML in infectious diseases management presents several challenges and limitations, which can be categorized into pathogen behaviour, model adaptability, and data management challenges.
First, the unpredictable and dynamic nature of infectious disease outbreaks, including varying transmission dynamics and impacts of interventions, makes modelling with ML complex. This complexity is primarily due to the multifaceted and often unknown variables associated with the emergence and spread of pathogens. Each infectious disease exhibits unique patterns in terms of its transmissibility, virulence, and response to environmental factors and control or preventive interventions. Specifically, the complexity of biological systems presents a significant challenge [Citation12–14]. Infectious diseases are influenced by a myriad of interconnected factors including genetic, environmental, and social variables. This complexity often exceeds the capacity of current ML models, which may not fully capture the non-linear interactions and emergent properties of biological systems. For instance, ML models struggle with the integration of multi-scale data ranging from molecular to epidemiological scales, which is crucial for understanding and predicting disease dynamics.
An example is the emergence of novel pathogens such as SARS-CoV-2 that introduces a high degree of uncertainty. Key characteristics like the incubation period, asymptomatic transmission, and mutation rates initially remain largely unknown, complicating predictive modelling efforts. Furthermore, the effectiveness of public health interventions, which can significantly influence the course of an outbreak, adds another layer of variability. These interventions are often subject to societal behaviours and policy decisions, making their impact challenging to quantify accurately in predictive models [Citation15]. Another example is the Ebola outbreak in West Africa between 2014 and 2016, which posed significant challenges in predictive modelling due to its complex transmission dynamics and varying factors [Citation16]. The sub-exponential growth patterns of Ebola, influenced by factors like social contact networks and cultural practices, differed significantly from diseases with aerosol transmission like influenza, complicating early modelling efforts. Response strategies, such as increasing Ebola Treatment Unit capacities and isolating patients, introduced additional variables that were difficult to incorporate accurately into models due to data limitations [Citation17]. Furthermore, the spatial and temporal analysis of the outbreaks requires understanding both local dynamics and cross-border transmission, challenged by the paucity of precise and timely data [Citation18].
Second, infectious agents like viruses and bacteria, can mutate rapidly, making it difficult for ML models to stay current without constant retraining. For example, the influenza virus, which is notorious for its high mutation rate undergoes antigenic drift each year, a process where small genetic changes accumulate over time. These changes can alter the virus’s surface proteins, hemagglutinin, and neuraminidase, enough that people’s immune systems may not recognize the virus, even if they were previously vaccinated or infected [Citation19]. As a result, ML models used to predict influenza trends or vaccine effectiveness need regular updates to incorporate the latest genetic information about circulating strains. Accordingly, systematically reviewing the literature is required to explore the use of ML techniques to generate predictions of influenza virus phenotypes based on genomic and/or proteomic datasets [Citation16]. The results showed that the ever-changing genetic landscape of the influenza virus poses a substantial challenge to applying ML in influenza management. The rapid evolutionary adaptability of pathogens like viruses adds another layer of difficulty [Citation20]. These organisms can mutate rapidly, sometimes even within the course of an outbreak, thereby altering their behaviour and confounding existing predictive models. This necessitates continuous updates to the models, a process that is both data-intensive and computationally demanding. The constant evolution of pathogens means that historical data might quickly become outdated, reducing the predictive accuracy of ML models unless they are frequently recalibrated with new data.
Third, ML encounters challenges in managing infectious diseases due to data issues. These include data availability for emerging pathogens, inconsistencies in records leading to biases, and variations across regions. These challenges pose a risk of perpetuating healthcare disparities. When it comes to real-time data integration for outbreaks, privacy concerns arise necessitating the implementation of anonymization techniques and governance protocols. The deployment of ML in clinical decision support systems (CDSS) requires a range of diverse data for training and validation purposes, including both structured and unstructured information [Citation5]. Ensuring the inclusivity of data and compatibility with different technology platforms is crucial for disease management. It is essential to address biases, particularly those affecting groups, while also ensuring that clinicians can properly interpret the outputs generated by ML-CDSS systems. Robust plans must be implemented to handle failures and cyber threats, along with validation processes to ensure safety and reliability in public health [Citation5].
In addition to data quality and interpretability challenges in ML for diseases, black-box models may present significant limitations [Citation21]. These models often lack transparency in their outputs like diagnostics or treatment plans, hindering understanding by data scientists and healthcare professionals. This obscurity can lead to misinterpretation or bias oversight, crucial for informed medical decisions.
Lastly, the socio-economic context plays a critical role in the spread and management of infectious diseases, and its variability across different regions and populations introduces additional complexity to modelling efforts [Citation22,Citation23]. Factors such as population density, healthcare infrastructure, public health policies, and community norms can significantly influence disease transmission and the effectiveness of intervention strategies. The ML models often do not adequately account for these diverse and dynamic social determinants of health, leading to potential biases and inaccuracy in predictions.
Opportunities to overcome ML limitations in infectious diseases management
Several review articles have explored the efficacy and limitations of ML techniques in managing infectious diseases. Santangelo et al. [Citation12] demonstrated the potential of ML to predict infectious disease outbreaks by combining various techniques to achieve accurate and plausible results. The study emphasized the need for high-quality data to enhance prediction capabilities. Another study provided an overview of how supervised ML techniques are increasingly applied in laboratory medicine for diagnosing various infectious diseases such as COVID-19, sepsis, and tuberculosis, and stressed the utility of these techniques in improving diagnostic accuracy and efficiency [Citation24]. A systematic review and meta-analysis analyzed the efficacy of ML models in predicting sepsis onset in intensive care units [Citation25]. Their work detailed how models like Random Forest and XGBoost can use clinical predictors such as age, creatinine levels, and vital signs to forecast sepsis onset, thereby potentially improving patient outcomes through early intervention.
To deal with the challenges posed by nature of pathogens and its outbreaks, it is crucial to develop adaptive models that can learn and update as data becomes available. An example of these models is a study by Bhadriraju and colleagues [Citation26], which introduced an adaptive model identification framework that uses sparse regression and feature selection to understand and predict the dynamics of complex processes with smaller data sets. They proposed a three-step procedure involving Sparse Identification of Nonlinear Dynamics (SINDy), ordinary least-squares regression and stepwise regression, demonstrating its effectiveness by comparing the modelling of a continuous stirred tank reactor with traditional SINDy methods [Citation26].
Additionally, using domain knowledge to incorporate information about similar pathogens is a promising strategy. A recent study developed a mixed-effect ML model to predict gene editing CRISPR interference (CRISPRi) guide efficiency in Escherichia coli using integrated data from multiple gene target screens [Citation27]. The model, which combines a linear random-effect model with a fixed-effect random forest, effectively separates the influence of the targeted gene from guide efficiency. The method applied in their work is insightful in situations where direct measurements are challenging to obtain.
Furthermore, ML models should be flexible to account for varying transmission patterns and intervention scenarios. A team of researchers from the United States developed a method that combines a Bayesian time series model and a random forest algorithm within a compartmental framework [Citation28]. This approach provided data-driven predictions for COVID-19. They evaluated the model by extending the training period throughout the pandemic and assessing its accuracy in forecasting over 21-day periods. Interestingly, the model revealed variations in projection trajectories and uncertainties across states, as observed in New York, Colorado, and West Virginia [Citation28].
To address the challenges posed by mutating agents and the lack of transparency in "black box" models used for disease prediction, various advanced methodologies are employed. Firstly, continuous learning approaches play a role as they allow models to automatically integrate data, ensuring that predictions remain relevant even with ever-changing pathogens [Citation29,Citation30]. This is particularly important when dealing with viruses that mutate quickly since traditional static models become outdated rapidly. Secondly, explainable AI (XAI) techniques are used to enhance transparency in model decision-making processes. The XAI enables an understanding of how models arrive at their decisions, which helps build trust and identify biases in algorithms [Citation30]. It plays a role in unravelling AI processes, making them accessible to a broader range of users, including healthcare professionals and policymakers. The main hurdle and significant challenges lie in harmonizing advancements with the practical aspects of healthcare, protecting data privacy, securing consent, and ensuring access to these advanced tools.
Soon, we envision progress in the field moving towards integrated and adaptable systems. There will likely be a transition towards developing models that are not only accurate but also understandable and transparent. This shift will foster trust and enhance usability among healthcare professionals. Additionally, we expect advancements in real-time data analysis and continuous learning models, enabling responses to emerging infectious diseases.
An area of research that currently holds intrigue is the use of ML in personalised medicine within the field of infectious diseases. This approach tailors treatment and prevention strategies based on patient profiles, potentially leading to better outcomes in infectious disease management. The incorporation of data with ML algorithms presents an avenue for developing such personalized strategies. However, it is imperative to address the logistical challenges associated with handling sensitive data.
Potential applications
Catastrophic evolution: biomolecular targets to reduce risks for infectious diseases
Catastrophic selection is a phenomenon associated with organisms resistant to pathogens and other stringent environments due to some deviant genome signatures and physiological differences [Citation31,Citation32]. Transposable sequences of viral origin or endogenous retroviruses (ERVs) are dormant in the modern human genome and when activated may regulate transcription contributing to innate immunity, cellular senescence, and tissue aging [Citation33,Citation34]. The hypothesis and recent evidence support that humans evolved with catastrophic selection associated with pathogen infection, which resulted in genomic signatures leading to biomolecular targets (proteins and post-translational modifications) to reduce risks for infectious diseases [Citation35]. To address this hypothesis, it is possible to apply comparative genomics to study human genome in comparison with Old-World monkeys and other species to identify inactivated genes that may be related to evolutionary adaptations to pathogen infection. For example, the major histocompatibility complex (MHC) is a chromosome 6 locus containing polymorphic genes that code for cell surface proteins essential for adaptive immunity. Comparative genomic analysis of MHC and other regions between sympatric host species might shed light on the adaptive immune system dynamics. Using ML algorithms and disease modelling it may be possible to find driving factors for host-pathogen interactions and disease susceptibility. Then, using a systems biology approach for analysis of omics datasets (transcriptomics, proteomics, metabolomics, interactomics, regulomics) to characterize molecular pathways and biomolecules associated with the identified inactivated genes, and applying ML algorithms to predict which identified biomolecular targets are able to interfere with pathogen infection from a biomedical perspective may result in candidate biomolecules for possible interventions to reduce risks for infectious diseases. Finally, considering that biomolecules do not act in isolation but interacting with each other forming a network collectively known as interactome, sub-networks of genes and proteins that interfere with pathogen infection are prime candidates for therapeutic target selection. For this, we may use both local and global network topological properties of genes/proteins such as degree and betweenness centrality as input features and explore graph representation learning with measures of success including interference with immune response pathways, metabolic pathways, and known action of pharmaceuticals for the prevention and control of infectious diseases. However, possible negative trade-off effects of biomolecules associated with catastrophic selection in some individuals should be considered and included in ML algorithms [Citation31].
Vaccinomics: high throughput discovery and characterization of vaccine protective antigens using intelligent Big Data analytics techniques
Vaccinomics is defined as the use of genome-scale omics technologies together with bioinformatics to have a holist approach for the development of next-generation vaccines [Citation36–38]. Regarding the use of AI and ML algorithms, one of the most promising applications is the prediction of vaccine efficacy and effectiveness based on the identification of the best protective antigens [Citation38]. However, the main obstacles include the lack of reliability on ML to use these predictive models on complex real cases and data composed of difficult to obtain features. The lack of reliability is based on two inherent aspects of this technology that increases with the more high-dimensional the data are. Although ML models have been validated and there are various metrics we can use, there is not an individual measure of certainty for each output. Some of the most used metrics are Mean Absolute Error (MAE), True/False negative/positive rates and derived metrics as the Area Under the Receiver Operating Characteristic curve (AUC-ROC CURVE), the recall or the precision. We must use these different indicators and interpret them in order to determine the quality of the model, but we cannot determine the interval of confidence of each individual prediction [Citation39]. Another problem is understanding the processes or decisions made by the algorithm to get the predictions [Citation40]. Second, the immunological datasets are based on laboratory experimentation and as previously mentioned the effect of evolutionary mechanisms make ML datasets difficult to keep updated due to modifications in pathogen and vector derived antigens and associated host adaptations thus compromising the flexibility of the model. Approaching these limitations requires the combination of different algorithms and Big Data analytics such as the combination of a supervised ML model as Random Forest (RF) with other non-supervised models such as Hierarchical Clustering (HCA) and Fuzzy Deformable Prototypes (FDP) to overcome the confidence and data complexity problems [Citation38]. The HC has been used as a step prior to RF training to reduce features in very complex and high dimensional datasets [Citation41,Citation42]. Additionally, and despite its limitations [Citation30], black-box models have been used to interpret the predictions of complex ML algorithms such as deducing the patterns learned by deep neural networks to understand how the algorithm works once trained and to detect biases [Citation43]. The HC could also be applied to make clusters of the different individual trees used to make the predictions and choose one of these trees to graphically represent the decision-making process in a simplified way and considering not only the predicted variable but also the variables used as inputs [Citation44]. The identification of vaccine antigens is a good example of FDP as there is a complex question in which it is difficult to choose a perfect prototype for a vaccine candidate considering that it is defined not as an element but as a group of good, mediocre, and bad elements of a category, being less intuitive but more accurate to reality [Citation45]. Accordingly, FDP could be used to facilitate prediction of antigen feasibility as a vaccine candidate and to extend the response variables predicted by the supervised method to obtain a more complex profile of the features of predicted interest. In summary, the combination of ML algorithms may lead to a robust Big Data method that allows us to predict the performance of antigens as vaccine candidates and considering data uncertainty and vagueness.
Additional applications. The potential applications discussed above are examples of ML approaches to infectious diseases. However, other potential applications including RNA interference-mediated gene control [Citation46], structure-based drug design [Citation47], and therapeutic interventions against drug resistance [Citation48] are under development with possible impact on the prevention and control of infectious and non-infectious diseases.
Conclusions
To advance in ML and AI in infectious diseases it is important to approach challenges with innovative solutions (). The application of ML for the management of infectious diseases holds promise but also faces significant challenges, such as the rapid mutation of pathogens, biases in data, and concerns about privacy. To overcome these challenges, it is crucial to adopt learning approaches, use interpretable models, and focus on inclusive data practices. Future directions include applications of ML to identify biomolecules for effective treatment and prevention of infectious diseases.
Authors contributions
Authors A.Z. Al Meslamani and J. de la Fuente: conception and design. Authors A.Z. Al Meslamani. I. Sobrino and J. de la Fuente: analysis and interpretation of the data. Authors A.Z. Al Meslamani. I. Sobrino and J. de la Fuente: drafting of the paper. Author J. de la Fuente: revising it critically for intellectual content; and the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Acknowledgments
The authors thank Margarita Villar and Marinela Contreras (Genomics, Proteomics and Biotechnology at SaBio, IREC), Agustin Estrada-Peña (University of Zaragoza) and José A. Olivas (Technologies and Information Systems Institute, University of Castilla-La Mancha) for their contribution to our initiatives on ML applications.
Disclosure statement
The author J. de la Fuente is section editor in the same journal. We declare no other competing interests.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Additional information
Funding
References
- Al Meslamani AZ, Jarab AS, Ghattas MA. The role of machine learning in healthcare responses to pandemics: maximizing benefits and filling gaps. J Med Econ. 2023;26(1):1–10. doi:10.1080/13696998.2023.2224018.
- Abou Hajal A, Al Meslamani AZ. Overcoming barriers to machine learning applications in toxicity prediction. Expert Opin Drug Metab Toxicol. 2023:1–5. doi:10.1080/17425255.2023.2294939.
- Park DJ, Park MW, Lee H, et al. Development of machine learning model for diagnostic disease prediction based on laboratory tests. Sci Rep. 2021;11(1):7567. doi:10.1038/s41598-021-87171-5.
- Zoabi Y, Kehat O, Lahav D, et al. Predicting bloodstream infection outcome using machine learning. Sci Rep. 2021;11(1):20101. doi:10.1038/s41598-021-99105-2.
- Peiffer-Smadja N, Rawson TM, Ahmad R, et al. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin Microbiol Infect. 2020;26(8):1118. doi:10.1016/j.cmi.2019.09.009.
- Yang J, Soltan AAS, Clifton DA. Machine learning generalizability across healthcare settings: insights from multi-site COVID-19 screening. NPJ Digit Med. 2022;5(1):69. doi:10.1038/s41746-022-00614-9.
- Kalipe G, Gautham V, Behera RK. Predicting malarial outbreak using machine learning and deep learning approach: a review and analysis. In Proceedings of the International Conference on Information Technology, ICIT. 2018; pp. 33–38. doi:10.1109/ICIT.2018.00019.
- Sebastianelli A, Spiller D, Carmo R, et al. A reproducible ensemble machine learning approach to forecast dengue outbreaks. Sci Rep. 2024;14(1):3807. doi:10.1038/s41598-024-52796-9.
- Abdulkareem AB, Sani NS, Sahran S, et al. Predicting COVID-19 based on environmental factors with machine learning. Intell Autom Soft Comput. 2021;28(2):305–320. doi:10.32604/iasc.2021.015413.
- Amin S, Uddin MI, Alsaeed DH, et al. Early detection of seasonal outbreaks from twitter data using machine learning approaches. Complexity. 2021;2021:1–12. doi:10.1155/2021/5520366.
- Moulaei K, Shanbehzadeh M, Mohammadi-Taghiabad Z, et al. Comparing machine learning algorithms for predicting COVID-19 mortality. BMC Med Inform Decis Mak. 2023;22(1):1–12. doi:10.1186/S12911-021-01742-0.
- Santangelo OE, Gentile V, Pizzo S, et al. Machine learning and prediction of infectious diseases: a systematic review. MAKE. 2023;5(1):175–198. doi:10.3390/make5010013.
- Erbe R, Gore J, Gemmill K, et al. The use of machine learning to discover regulatory networks controlling biological systems. Mol Cell. 2022;82(2):260–273. doi:10.1016/j.molcel.2021.12.011.
- Zhao AP, Li S, Cao Z, et al. AI for science: predicting infectious diseases. J Safety Sci Resilience. 2024;5:130–146. doi:10.1016/j.jnlssr.2024.02.002.
- Holmdahl I, Buckee C. Wrong but useful - What Covid-19 epidemiologic models can and cannot tell us. N Engl J Med. 2020;383(4):303–305. doi:10.1056/NEJMp2016822.
- Chowell G, Viboud C, Simonsen L, et al. Perspectives on model forecasts of the 2014–2015 Ebola epidemic in West Africa: lessons and the way forward. BMC Med. 2017;15(1):42. doi:10.1186/s12916-017-0811-y.
- Meltzer MI, Santibanez S, Fischer LS, et al. Modeling in real time during the Ebola response. Morbidity and Mortality Weekly Report are service marks of the U.S. Department of Health and Human Services. 2016 [cited 2024 Jan 10]. https://www.cdc.gov/mmwr/volumes/65/su/su6503a12.htm#suggestedcitation.
- Backer JA, Wallinga J. Spatiotemporal analysis of the 2014 Ebola epidemic in West Africa. PLoS Comput Biol. 2016;12(12):e1005210. doi:10.1371/journal.pcbi.1005210.
- Borkenhagen LK, Allen MW, Runstadler JA. Influenza virus genotype to phenotype predictions through machine learning: a systematic review. Emerg. Microbes Infect. 2021;10(1):1896–1907. doi:10.1080/22221751.2021.1978824.
- Shi A, Fan F, Broach JR. Microbial adaptive evolution. J Ind Microbiol Biotechnol. 2022;49(2):kuab076. doi:10.1093/jimb/kuab076.
- Giacobbe DR, Zhang Y, de la Fuente J. Explainable artificial intelligence and machine learning: novel approaches to face infectious diseases challenges. Ann Med. 2023;55(2):2286336. doi:10.1080/07853890.2023.2286336.
- Feldmeyer D, Meisch C, Sauter H, et al. Using OpenStreetMap data and machine learning to generate socio-economic indicators. IJGI. 2020;9(9):498. doi:10.3390/ijgi9090498.
- Kondeti PK, Ravi K, Mutheneni SR, et al. Applications of machine learning techniques to predict filariasis using socio-economic factors. Epidemiol Infect. 2019;147:e260. doi:10.1017/S0950268819001481.
- Tran NK, Albahra S, May L, et al. Evolving applications of artificial intelligence and machine learning in infectious diseases testing. Clin Chem. 2021;68(1):125–133. doi:10.1093/CLINCHEM/HVAB239.
- Yang Z, Cui X, Song Z. Predicting sepsis onset in ICU using machine learning models: a systematic review and meta-analysis. BMC Infect Dis. 2023;23(1):635. doi:10.1186/S12879-023-08614-0/FIGURES/13.
- Bhadriraju B, Narasingam A, Kwon JS-I. Machine learning-based adaptive model identification of systems: application to a chemical process. Chem Eng Res Des. 2019;152:372–383. https://www.sciencedirect.com/science/article/pii/S0263876219304162 doi:10.1016/j.cherd.2019.09.009.
- Yu Y, Gawlitt S, de Andrade e Sousa LB, et al. Improved prediction of bacterial CRISPRi guide efficiency from depletion screens through mixed-effect machine learning and data integration. Genome Biol. 2024;25(1):13. doi:10.1186/s13059-023-03153-y.
- Watson GL, Xiong D, Zhang L, et al. Pandemic velocity: forecasting COVID-19 in the US with a machine learning & Bayesian time series compartmental model. PLoS Comput Biol. 2021;17(3):e1008837. doi:10.1371/journal.pcbi.1008837.
- Tomašev N, Harris N, Baur S, et al. Use of deep learning to develop continuous-risk models for adverse event prediction from electronic health records. Nat Protoc. 2021;16(6):2765–2787. doi:10.1038/s41596-021-00513-5.
- Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019;572(7767):116–119. doi:10.1038/s41586-019-1390-1.
- Galili U. Evolution in primates by "Catastrophic-selection" interplay between enveloped virus epidemics, mutated genes of enzymes synthesizing carbohydrate antigens, and natural anti-carbohydrate antibodies. Am J Phys Anthropol. 2019;168(2):352–363. doi:10.1002/ajpa.23745.
- Lewis H. Catastrophic selection as a factor in speciation. Evolution. 1962;16:257–271. doi:10.2307/2406275.
- Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351(6277):1083–1087. doi:10.1126/science.aad5497.
- Liu X, Liu Z, Wu Z, et al. Resurrection of endogenous retroviruses during aging reinforces senescence. Cell. 2023;186(2):287–304.e26. doi:10.1016/j.cell.2022.12.017.
- Maasch JRMA, Torres MDT, Melo MCR, et al. Molecular de-extinction of ancient antimicrobial peptides enabled by machine learning. Cell Host Microbe. 2023;31(8):1260–1274.e6. doi:10.1016/j.chom.2023.07.001.
- Poland GA, Kennedy RB, McKinney BA, et al. Vaccinomics, adversomics, and the immune response network theory: individualized vaccinology in the 21st century. Semin Immunol. 2013;25(2):89–103. doi:10.1016/j.smim.2013.04.007.
- de la Fuente J, Merino O. Vaccinomics, the new road to tick vaccines. Vaccine. 2013;31(50):5923–5929. doi:10.1016/j.vaccine.2013.10.049.
- de la Fuente J, Villar M, Estrada-Peña A, et al. High throughput discovery and characterization of tick and pathogen vaccine protective antigens using vaccinomics with intelligent Big Data analytic techniques. Expert Rev Vaccines. 2018;17(7):569–576. doi:10.1080/14760584.2018.1493928.
- Coulston JW, Blinn CE, Thomas VA, et al. Approximating prediction uncertainty for random Forest regression models. Photogram Engng Rem Sens. 2016;82(3):189–197. doi:10.14358/PERS.82.3.189.
- Ribeiro MT, Singh S, Guestrin C. Model-agnostic interpretability of machine learning. In Presented at 2016 ICML Workshop on Human Interpretability in Machine Learning (WHI 2016), New York, NY [cited 2024 Feb 23]. https://arxiv.org/abs/1606.05386v1.
- Liu H, Wu X, Zhang S. Feature selection using hierarchical feature clustering. International Conference on Information and Knowledge Management, Proceedings. 2011 [cited 2024 Feb 23];979–984. https://dl.acm.org/doi/10.1145/2063576.2063716. doi:10.1145/2063576.2063716.
- Alizadeh-Sani Z, Martínez PP, González GH, et al. A hybrid supervised/unsupervised machine learning approach to classify web services. Commun Comput Inform Sci. 2021;1472:93–103. https://link.springer.com/chapter/10.1007/978-3-030-85710-3_8 doi:10.1007/978-3-030-85710-3_8.
- Singh C, Yu B, James Murdoch W. Hierarchical interpretations for neural network predictions. In 7th International Conference on Learning Representations, ICLR 2019. 2018 Jun 14 [cited 2024 Feb 23]. https://arxiv.org/abs/1806.05337v2.
- Chen D, Goyal G, Go RS, et al. Improved interpretability of machine learning model using unsupervised clustering: predicting time to first treatment in chronic lymphocytic leukemia. J Clin Oncol Clin Cancer Inform. 2019;3:1–11. https://ascopubs.org/doi/10.1200/CCI.18.00137 doi:10.1200/CCI.18.00137.
- Olivas Varela JÁ. Contribución al estudio experimental de la predicción basada en categorías deformables borrosas. 2000 [cited 2024 Feb 23]. https://dialnet.unirioja.es/servlet/tesis?codigo=8741&info=resumen&idioma=SPA.
- Aljedaie MM, Alam P. In silico identification of human microRNAs pointing centrin genes in Leishmania donovani: considering the RNAi-mediated gene control. Front Genet. 2024;14:1329339. doi:10.3389/fgene.2023.1329339.
- Sulea T, Kumar S, Kuroda D. Editorial: progress and challenges in computational structure-based design and development of biologic drugs. Front Mol Biosci. 2024;11:1360267. doi:10.3389/fmolb.2024.1360267.
- Gopikrishnan M, Haryini S, C GPD. Emerging strategies and therapeutic innovations for combating drug resistance in Staphylococcus aureus strains: a comprehensive review. J Basic Microbiol. 2024;64(5):e2300579. doi:10.1002/jobm.202300579.