Abstract
Our aim was to find out differences between chronic periodontitis (CP) and periodontally healthy subjects with respect to oral lactic acid bacteria (LAB) as well as subgingival microbial relations. Clinical data, salivary levels of lactobacilli and mutans streptococci, and subgingival microbial samples were obtained from 26 CP and 15 periodontally healthy subjects. Antimicrobial activity of LAB against periodontal pathogens was assessed. We found Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Prevotella intermedia/nigrescens in 54%, 23% and 73% of CP patients, respectively; the latter was also found in 47% of healthy subjects. The mean proportion of streptococci (27.0 vs 15.2%), particularly S. mutans group (5.7 vs 0.8%) and S. mitis group (18.1 vs 5.0%), as well as aerobic coryneforms (24.7 vs 11.9%) was higher in healthy persons (p < 0.05). An inverse relationship of subgingival streptococci and aerobic coryneforms with periodontal pathogens and deteriorated clinical parameters were seen. Salivary counts of mutans streptococci were higher in healthy persons. Inhibition of periodontal pathogens by LAB was observed. In conclusion, the proportions of oral LAB were significantly lower in CP than in healthy subjects, whilst being important antagonists against periodontal pathogens. These oral commensals may play an important role in the suppression of periodontal pathogens and maintenance of microecological balance in the oral cavity.
Introduction
Periodontitis is a chronic inflammation of the periodontium that results in periodontal tissue destruction and alveolar bone loss. Tissue destruction occurs as a consequence of the host's attempt to eliminate bacteria from the gingival sulcus by evoking an immuno-inflammatory response. A report from the World Workshop in Periodontics in 1996 emphasized the polymicrobial nature of chronic periodontitis, but most frequently it has been related to the black-pigmented gram-negative anaerobic rods Porphyromonas gingivalis and Prevotella intermedia, accompanied by the gram-negative microaerobic coccobacillus Actinobacillus actinomycetemcomitansCitation[1].
Although there have been several studies on putative periodontal pathogens, less is known about the microorganisms that might participate in the maintenance of periodontal health. There are numerous microbiotopes in the mouth that are colonized by a variety of microorganisms playing an active role in the maintenance of local oral health as well as influencing each other through various synergistic and antagonistic interactions. Some oral commensals such as lactic acid bacteria (LAB), including among others the genera Streptococcus and Lactobacillus, may prohibit periodontal pathogens. Hillman et al. Citation[2] showed that the presence of the putative periodontal pathogens P. gingivalis, P. intermedia and A. actinomycetemcomitans in subgingival plaque of periodontitis patients was correlated with the absence of certain streptococci (e.g. Streptococcus sanguis). Investigating mutans streptococci in the subgingival plaque of periodontitis patients at different stages of periodontal therapy, van der Reijden et al. Citation[3] found their proportions to increase following therapy, as well as a negative correlation between mutans streptococci and P. gingivalis. However, they did not relate the microbiological data to the clinical measurements.
Recent studies have also described the ability of streptococci and lactobacilli to exert antibacterial activity against various pathogens, including P. gingivalis and P. intermediaCitation[4], Citation[5]. Thus, we may suppose that colonization of the oral cavity by LAB may aid in suppression of periodontal pathogens and hence reduce the incidence of chronic periodontitis (CP). However, the existing data do not provide a clear picture as to whether colonization with LAB differs with respect to periodontal health and how the microecological relations between LAB and other microorganisms are changed in CP patients and periodontally healthy subjects.
The objective of the present study was to find out differences in the colonization of LAB in saliva and periodontal sites in CP and periodontally healthy subjects. In addition, we aimed to investigate subgingival microbial relations with respect to different clinical parameters.
Materials and methods
Study group
The study group included 26 patients with CP (16 female, 10 male, mean age 47.2±11.3 years) and 15 periodontally healthy subjects (7 female, 8 male, mean age 37.5±10.4 years); both groups had no history of systemic disease or antibiotic therapy within the 6 months prior to sampling. The CP patients were diagnosed as having CP based on gingival inflammation, periodontal breakdown with pocket depth ≥5 mm and radiographic evidence of bone loss Citation[6]. They were consecutively drawn from the waiting list of patients who were referred to the Department of Oral Surgery at the Clinic of Stomatology of Tartu University for diagnosis and treatment of periodontitis during the years 1999–2001. Healthy individuals were defined as having no radiographic or clinical evidence of attachment loss. The smoking history of the subjects was checked according to a questionnaire. In the periodontitis group 9 patients were non-smokers, 2 were current smokers and 15 were former smokers who had stopped smoking on average 16.1 (±7.5) years ago. In the healthy group 13 subjects were non-smokers and 2 were former smokers who had stopped smoking on average 3.0 (±1.4) years ago.
All patients were screened for their suitability and selection of sampling sites a day prior to collection of microbiological samples.
Informed consent was obtained from all subjects, in accordance with the procedures of the Ethics Review Committee on Human Research of the University of Tartu.
Clinical examination
The baseline examination included the registration of dental plaque, gingival inflammation and presence of suppuration on probing at four sites (distal, mesial, lingual and buccal sites), and periodontal probing depth and attachment level at six sites (distal, mid and mesial aspects for both buccal and lingual sites) of each tooth, excluding third molars.
The plaque status of an individual was given as the plaque index (PI) according to Silness and Löe Citation[7] and as the frequency of plaque-positive surfaces expressed as a percentage of the total number of surfaces. Gingival inflammation was given as a modified gingival index (GI: 0, healthy gingiva with no bleeding on probing; 1, ‘pinprick’ bleeding on probing; 2, immediate and overt bleeding on probing; 3, spontaneous bleeding) Citation[8], Citation[9] and as the frequency of bleeding sites on probing, expressed as a percentage of all sites. Suppuration on probing was recorded as present or absent and the results were given as the frequency of pus-positive sites expressed as a percentage of all sites. Periodontal probing depth (PPD) was measured to the nearest millimetre from the gingival margin to the bottom of the gingival sulcus/pocket and periodontal attachment level (PAL) from the cemento-enamel junction to the bottom of the periodontal pocket with a Williams periodontal probe. The mean across all sites formed the PPD and attachment level of the patient, designated as PPDall and PALall, respectively. Sites with a probing depth of ≥5 mm were defined as diseased sites (DS). The frequency of DS was expressed as percentage of the total number of sites. The mean PPD and attachment level of diseased sites was designated as PPDds, and PALds, respectively. In addition, dental caries was registered in accordance with the WHO criteria Citation[10]. The same examiner performed all clinical measurements.
Salivary levels of lactobacilli and mutans streptococci
Salivary lactobacilli were investigated in 20 CP patients and 15 healthy subjects by the Dentocult®LB dip-slide method (Orion Diagnostica, Espoo, Finland), and mutans streptococci in 14 CP and 14 healthy patients by the Dentocult®SM strip method (Orion Diagnostica) in paraffin-stimulated saliva Citation[11]. After incubation for 3 days at 37°C the number of lactobacilli and mutans streptococci per ml of saliva was estimated by comparing the slides with a density chart provided by the manufacturer. Dentocult®LB test results were expressed as missing, low (≤103 CFU/ml), medium (104 CFU/ml), high (105 CFU/ml) and very high (>106 CFU/ml) counts of lactobacilli. Dentocult®SM test results were given as missing, low (<105 CFU/ml), medium (105–106 CFU/ml) and high (>106 CFU/ml) counts of mutans streptococci.
Subgingival samples
In periodontitis patients the two deepest periodontal pockets with inflammation, one in the upper and one in the lower jaw, were selected for sampling (e.g. the first sample was taken from the deepest pocket with inflammation in the first quadrant and the second sample was taken from the deepest pocket with inflammation in the third quadrant). In total, 54% of sampled sites in periodontitis patients were located in the molar/premolar region and 46% in the canine/incisor region. In healthy subjects two clinically healthy gingival sulci, one in the molar/premolar region and the other in the canine/incisor region in opposite jaws, were selected for sampling. The samples were obtained by means of a gingival crevice lavage method as described by Boström et al. Citation[12]. Prior to sampling, the area was isolated with cotton rolls and the supragingival region of the tooth surface to be sampled was cleaned and dried with sterile cotton pellets. A reduced transport fluid (RTF) was used as the sampling fluid. Small volumes (10–20 µl) were ejected from a glass ampoule with a cannula into the periodontal pocket about 1 mm from the bottom and aspirated into the ampoule. The total volume of sampling fluid was 250 µl. The ejection and aspiration procedure was repeated four times. Four drops (about 40 µl) were transferred from the ampoule to a vial containing anaerobic transport medium, VMGA III. The samples were processed within 2 h.
Microbiological analysis
After vortex mixing for 30 s, samples were 10-fold serially diluted in pre-reduced peptone water (Oxoid, Unipath, Basingstoke, UK) and 100 µl of appropriate dilutions were plated onto agar media. The following media were used: Brucella agar (Oxoid), supplemented with 5% defibrinated horse blood and menadion (2.5 µg/ml) Citation[13] for enumeration of anaerobic and facultative anaerobic bacteria; tryptone soya agar (Oxoid), supplemented with yeast extract (0.1%), horse serum (10%), bacitracin (75 µg/ml) and vancomycin (5 µg/ml) (TSBV) for identification of A. actinomycetemcomitans Citation[14]; and MRS agar (Oxoid) for lactobacilli and streptococci. Brucella plates were incubated in an anaerobic glove box (Sheldon Manufacturing, Inc., Shel LAB, Cornelius, OR, USA) with a gas mixture of 5% H2, 5% CO2, 90% N2 for 5–6 days. TSBV and MRS plates were incubated in a microaerophilic atmosphere (10% CO2) for 72 h.
Colonies with different morphology were Gram stained and examined microscopically. The microorganisms were identified mostly to the genus level by standard methods Citation[15]. Streptococci and enterococci were identified by the absence of catalase production and differentiated by the fermentation of esculin in the presence of bile. Viridans streptococci were distinguished from Streptococcus pneumoniae by the optochin susceptibility test and from Streptococcus bovis by the bile esculin test, and were further grouped by hydrolysis of arginine and production of acetoin (Voges-Proskauer test). Gram-positive rod-shaped non-spore-forming cells expressing a negative catalase reaction and multiplying in microaerobic conditions on MRS medium were considered as Lactobacillus species and were further identified by molecular methods Citation[16]. A. actinomycetemcomitans was differentiated by colony morphology (star-like inner structure) on selective medium, cell morphology and catalase production, inability to ferment sucrose and absence of beta-glucuronidase activity using medium containing MUG supplement (Oxoid).
The anaerobes were identified by their colony and cellular morphology and Gram stain reaction and some rapid tests and diagnostic disks (catalase, oxidase, spot indole, fluorescence, oxgall, brilliant green, bile esculin, colistin, vancomycin, kanamycin). Gram-positive anaerobic non-spore-forming rods with negative catalase and indole reaction were identified as Eubacterium spp. and spore-forming bacteria as Clostridium spp. Gram-positive irregularly shaped non-spore-forming anaerobic or facultatively anaerobic rods were classified respectively as ‘anaerobic coryneforms’ and ‘aerobic coryneforms’ and included species of Actinomyces, Corynebacterium, Propionibacterium and Bifidobacterium. Gram-negative anaerobic rods that formed black-pigmented colonies, were vancomycin-resistant, colistin-sensitive, indole-positive and showed brick-red fluorescence under UV light, were presumptively identified as P. intermedia/nigrescens, and black-pigmented colonies with vancomycin sensitivity, colistin resistance, indole production and no fluorescence as P. gingivalis. All anaerobic microorganisms were tested for absence of growth under aerobic and microaerophilic conditions on freshly prepared chocolate agar and blood agar plates.
In vitro antimicrobial activity testing
Antimicrobial activity of Streptococcus mutans NG8 (wild type) against target bacteria A. actinomycetemcomitans 31-1-1A (wild type), A. actinomycetemcomitans 31-2-1A (wild type), P. gingivalis ATCC 49417, P. gingivalis W83 and P. intermedia ATCC 25611 was assessed using a streak line procedure Citation[17] on Wilkins-Chalgren blood agar plates (Oxoid). The same method was used to test antimicrobial activity of subgingival lactobacilli (three strains of L. gasseri, two of L. oris and one of L. paracasei) against P. gingivalis ATCC 49417 and P. intermedia ATCC 25611. A single line of S. mutans or lactobacilli culture (grown in MRS broth for 48 h at 37°C in a 10% CO2 environment) was seeded in the middle of the agar plate, and cultivated for 48 h at 37°C in an anaerobic glove box (Sheldon Manufacturing, Inc.) with a gas mixture of CO2/H2/N2 at 5%/5%/90%, respectively, for testing against anaerobic bacteria (P. gingivalis, P. intermedia) and in 10% CO2 for testing against A. actinomycetemcomitans. Target bacteria P. gingivalis and P. intermedia were cultured in Wilkins-Chalgren broth for 48 h at 37°C in anaerobic conditions and A. actinomycetemcomitans in tryptone soya broth, supplemented with yeast extract, in microaerophilic conditions for 48 h at 37°C. Thereafter, a 10 µl aliquot of target bacterial culture (2.0 on the McFarland turbidity scale) was streaked in duplicate perpendicular to the streak line of S. mutans or lactobacilli. Following incubation of the plates for 72 h at 37°C in anaerobic or microaerophilic conditions depending on the target bacteria used, the width of the zone of inhibition (mm) of the target bacteria extending from the culture line of S. mutans or lactobacilli was measured.
The streak line procedure Citation[17] was also used to assess the antimicrobial activity of the above-described periodontal pathogens against S. mutans NG8 and lactobacilli. The periodontal pathogens were precultivated in their appropriate media and incubation conditions for 48 h at 37°C, streaked in the middle of the Wilkins-Chalgren blood agar plate and incubated for 48 h at 37°C in anaerobic (P. gingivalis, P. intermedia) or microaerophilic (A. actinomycetemcomitans) conditions. The target bacterial strains S. mutans and lactobacilli were cultured on MRS agar for 48 h and 24 h, respectively, in microaerophilic conditions, suspended in saline solution (1.0 on the McFarland turbidity scale) and streaked (10 µl) in duplicate perpendicular to the streak line of test bacteria. Following incubation of the plates for 72 h at 37°C in anaerobic or microaerophilic conditions depending on the periodontal pathogens used, the width of the zone of inhibition (mm) of the target bacteria extending from the culture line of periodontal pathogens was measured.
Statistical methods
The total count (log10 CFU/ml – colony forming units per millilitre of crevicular fluid) of microorganisms and the counts of various genera and species were calculated for each patient. The detection level of the various microorganisms was 3 log10 CFU/ml. A proportion (%) of the particular microbe in the total count was estimated. Statistical analyses were performed using SigmaStat (Jandel Scientific) and Excel (Microsoft Corp.). The following tests were employed: Fisher exact test, t test and Mann-Whitney rank sum test (comparison of different study and bacterial groups), Pearson product moment correlation (measuring correlations between the proportion (%) of various subgingival bacteria and the clinical indices) and Spearman rank order correlation (measuring correlations between the proportions of various microbes).
Results
Clinical characteristics
The clinical parameters of the subjects are shown in . The mean number of teeth present was significantly lower in the periodontitis group. We could observe much higher occurrence of dental plaque, a significant increase in all measured inflammatory parameters and much higher mean PPD in the CP patients as compared with the healthy group. Approximately a quarter of all measured periodontal sites in CP patients were equal to or deeper than 5 mm. With respect to the experience of dental caries, no statistically significant differences were observed between the two groups.
Table I. Clinical parameters of chronic periodontitis (CP) patients and periodontally healthy individuals.
Salivary levels of lactobacilli and mutans streptococci
All healthy subjects and 18 of 20 CP patients harboured salivary lactobacilli. Their counts were similar – very high, high and medium count in 80% of healthy and 70% of CP patients, and missing and low count in 20% vs 30%, respectively.
Mutans streptococci were isolated from all subjects in both groups; however, a trend for an inverse relationship was observed: the counts were low in 64% of CP patients and 36% of healthy subjects, while medium and high counts were seen in 36% of CP and 64% of healthy subjects.
Subgingival microflora
Mixed aerobic and anaerobic subgingival microflora was seen in all studied subjects (). The total bacterial count in diseased patients was much higher than in healthy individuals. The typical periodontal pathogens A. actinomycetemcomitans and P. gingivalis were only found in individuals with CP. P. intermedia/nigrescens was isolated from both diseased and healthy individuals with higher prevalence and count in the former group. The mean number of periodontal pathogens was significantly higher in CP patients than in healthy subjects (1.50±0.95 vs 0.47±0.52, p = 0.001).
Table II. Composition of the subgingival microflora in chronic periodontitis (CP) patients and periodontally healthy subjects.
The counts of different gram-negative anaerobic rods, Streptococcus anginosus group, peptostreptococci and anaerobic coryneforms were significantly higher in diseased than in healthy individuals. The former were also somewhat less often colonized by a gram-positive rod, Eubacterium spp., as compared with periodontally healthy subjects.
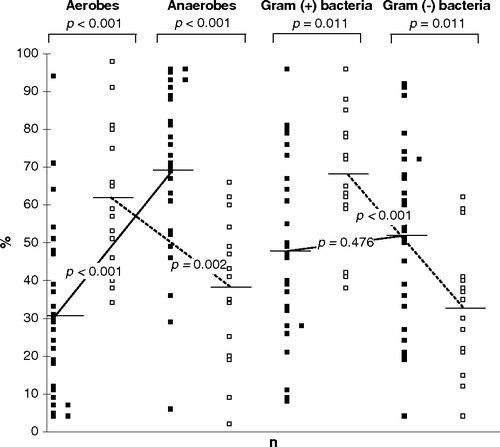
A strong inverse relationship in the proportion of aerobic and anaerobic as well as gram-positive and gram-negative microorganisms was found in periodontally diseased sites of CP patients and in healthy sites of healthy subjects (). The mean proportion of anaerobic bacteria, particularly gram-negative anaerobic rods (29.9±22.4% vs 12.7±11.4%, p < 0.001) and gram-positive anaerobic coryneforms (8.7±10.9% vs 2.5±3.7%, p < 0.001) was significantly higher in diseased sites, while the proportion of facultative anaerobic gram-positive bacteria – particularly streptococci (27.0±23.4% vs 15.2±19.2%, p = 0.012) and aerobic coryneforms (24.7±27.5% vs 11.9±14.7%, p = 0.028) – was significantly higher in healthy sites. Within the group of streptococci the mean proportion of S. mutans group (5.7±9.7% vs 0.8±1.8%, p = 0.015) and S. mitis group (18.1±18.9% vs 5.0±5.8%, p = 0.004) was significantly higher in healthy sites.
Figure 1. The proportion of aerobic, anaerobic, gram-positive and gram-negative bacteria expressed as a percentage of the total subgingival flora of chronic periodontitis (solid squares, n=26) patients and periodontally healthy (open squares, n=15) subjects. Each symbol represents a mean proportion of a particular microbial group in a subject. Perpendicular lines represent a mean value for each group of microorganisms averaged across subjects. All microaerophilic and facultatively anaerobic bacteria were included in the ‘aerobes’ and all obligately anaerobic bacteria in the ‘anaerobes’.
Positive correlations between the proportions of subgingival periodontal pathogens with each other and negative correlations with streptococci and aerobic coryneforms are shown in . A. actinomycetemcomitans and P. gingivalis were in stronger negative association with streptococci, and P. intermedia/nigrescens with coryneforms.
Table III. Correlations of the proportions of periodontal pathogens and clinical parameters with the proportions of streptococci, aerobic coryneforms and anaerobic gram-negative rods at respective subgingival sites.
Similar tendencies were observed when these microorganisms were correlated with CP-related clinical parameters. Plaque index (PI), gingival index (GI) and periodontal probing depth (PPD) were positively associated with the proportion of subgingival gram-negative anaerobic rods, and negatively with the proportion of subgingival streptococci and aerobic coryneforms ().
In vitro antimicrobial activity between lactic acid bacteria and periodontal pathogens
Inhibition of the periodontal pathogens A. actinomycetemcomitans, P. gingivalis and P. intermedia by S. mutans was observed (), with strongest antimicrobial activity towards P. gingivalis. Antimicrobial activity of lactobacilli against anaerobes was species-specific. All lactobacilli inhibited the growth of P. gingivalis, whereas only strains of L. gasseri and L. paracasei were able to inhibit P. intermedia.
Table IV. Antimicrobial activity of S. mutans and subgingival lactobacilli against putative periodontal pathogens.
On the other hand, none of the strains of A. actinomycetemcomitans and P. gingivalis were able to inhibit the growth of S. mutans, and only mild antimicrobial activity was expressed by P. intermedia (mean zone of inhibition 2.9±0.2 mm). Neither P. gingivalis nor P. intermedia inhibited the growth of lactobacilli.
Discussion
In the present study we found that the proportion of LAB was decreased in oral microflora in patients with CP as compared with healthy subjects.
Total counts of subgingival microorganisms
As expected and in agreement with previous reports Citation[1], Citation[18], the mean total count of microorganisms was much higher in periodontitis patients. This could be explained by different subgingival environmental conditions, because as revealed by clinical data, the periodontitis subjects had significantly deeper periodontal sites with obviously larger area for bacterial colonization. In addition, the high level of inflammation in diseased sites probably increased the amount of necessary nutrients for bacterial multiplication. Moreover, the possible mechanisms aiming to keep the bacterial numbers under control might have weakened. Large inter-individual variations in the number of different species and their counts were seen. This clearly supports the data on the individuality of a host's microflora, assessed in different biotopes of gastrointestinal and urogenital tracts Citation[19].
Periodontal pathogens
In our study, along with the increase in the absolute numbers of microbes, the number of suspected periodontal pathogens (P. gingivalis, P. intermedia/nigrescens, A. actinomycetemcomitans) also increased in periodontitis, yet the prevalence of the acknowledged periodontal pathogen P. gingivalis remained rather low, contradicting earlier reports Citation[9], Citation[20]. This could be explained by the differences in methodology (culturing vs molecular techniques), but recently the regional differences in the composition of the subgingival microbiota have been revealed as well Citation[21]. We also observed that periodontally healthy persons were never colonized with A. actinomyctemcomitans and P. gingivalis, while the colonization with P. intermedia/nigrescens was seen in half of them. These data suggest that the mere presence of a single periodontal pathogen does not necessarily lead to periodontal destruction. Instead, the possibility of causing inflammation may be dependent on the particular combination of various microorganisms Citation[20], Citation[22], as the mixed infection with two or three pathogens was seen in nearly half of the periodontitis patients. Also, the differences in the virulence of a particular strain might influence the outcome of the infection Citation[23].
Relationships between different groups of microorganisms
In addition to the usual analysis of the prevalence and total counts of species and genera of microbes, the proportions (%) of bacteria from the total count of microorganisms in a particular periodontal site were calculated. Due to the large individual variations in the absolute numbers of bacteria in different individuals Citation[19], we considered the proportions of microbes in the total count to be more informative. We found that anaerobic and gram-negative species predominated in the periodontitis patients, while aerobic and gram-positive species predominated in healthy individuals – confirming some earlier findings Citation[18], Citation[24]. The significant increase in the proportion of different anaerobic gram-negative rods in diseased sites occurred mostly at the expense of certain aerobic gram-positive bacteria, as periodontally healthy patients harboured much higher proportions of streptococci and aerobic coryneforms in their subgingival sites.
Lactic acid bacteria and periodontitis
The data from the present study show that colonization by LAB varies in different biotopes within the mouth. Lactobacilli, although frequently isolated from saliva, were seldom found from subgingival sites. Only 2 of 82 investigated subgingival sites were colonized by lactobacilli, which clearly shows that the subgingival region is not their common habitat. On the other hand, streptococci were frequently found in both saliva and subgingival sites, confirming earlier results Citation[3] and indicating that they could be the guardians aiding in the balance of subgingival microecology.
Streptococci and coryneforms (e.g. Actinomyces spp.) have frequently been found in healthy individuals Citation[18], Citation[22], Citation[24] and their possible beneficial role in subgingival plaque has been emphasized Citation[25]. In the present study we found a significantly higher proportion of streptococci, particularly S. mutans and S. mitis groups, as well as aerobic coryneforms among healthy persons. In addition, correlation analysis showed that a high proportion of these bacteria in a periodontal site was inversely associated with that of periodontal pathogens, suggesting antagonistic interactions between LAB and periodontal pathogens. Clinical investigations have shown that periodontal treatment significantly increases the proportions of streptococci (e.g. mutans streptococci) in both subgingival plaque and saliva Citation[3], Citation[26], confirming our results where both salivary and subgingival streptococci were in positive association with periodontal health.
Bacterial interactions, including antagonism, are likely to play an important role in the ecology of the microflora found in subgingival areas. Several inhibitory substances have been identified, including hydrogen peroxide, organic fatty acids, lactic acid, antibiotics, enzymes and bacteriocins. Inhibition of the in vitro growth of periodontal pathogens by viridans streptococci due to the production of hydrogen peroxide Citation[2] and antimicrobial activity of Actinomyces strains due to the production of various organic acids has been reported Citation[27]. Recently, Doran et al. Citation[5] showed that anaerobic bacteria (including P. intermedia and P. gingivalis) were inhibited by metabolic end products of glucose fermentation by oral streptococci, particularly strains of S. mutans and S. salivarius. We succeeded in confirming the ability of both S. mutans and lactobacilli to inhibit the growth of anaerobic P. gingivalis and P. intermedia by our in vitro antimicrobial activity testing. In addition, S. mutans suppressed microaerophilic A. actinomycetemcomitans, although to a lesser extent than anaerobic bacteria. As the pH is an important determinant of the microbial composition of plaque, the production of organic acids from carbohydrate fermentation by lactic acid-producing bacteria and concomitant fall in pH can interfere with the growth of surrounding microorganisms, including putative periodontal pathogens Citation[4], Citation[5]. The inability of P. gingivalis to grow at a pH below 6.5 Citation[28] makes its growth dependent on environmental conditions, and conceivably, the ecological determinants that reduce the proportion of LAB may favour the periodontal disease-inducing flora due to insufficient control mechanisms. Thus, the measures against decreasing the proportion of LAB microflora by antiseptics should be supported with evidence-based studies in different clinical situations, e.g. prevention of different oral diseases, not merely dental caries.
In summary, several microecological relations were substantially changed in CP patients as compared with periodontally healthy individuals, the major finding being a significant decrease in the proportions of oral streptococci and aerobic coryneforms in the former group. The data suggest that these oral commensals may play an important role in the suppression of periodontal pathogens and maintenance of microecological balance in the oral cavity. Thus, the excessive use of broad-spectrum antimicrobial agents against gram-positive microflora might shift the balance towards the overgrowth of potentially pathogenic species due to insufficient control mechanisms.
This study was supported by the Estonian Science Foundation (grant no. 5692). We thank Dr Lars E. Linder (Karolinska Institute, Sweden) for his valuable advice by starting this study, Eha-Maie Laanes (University of Tartu, Estonia) for her technical assistance and Docent Krista Fischer (University of Tartu, Estonia) for her valuable statistical advice. The bacterial strains S. mutans NG8, P. gingivalis W83, P. gingivalis ATCC 49417 and P. intermedia ATCC 25611 were kindly provided by Harold Marcotte (Karolinska Institute, Sweden).
References
- Zambon JJ. Periodontal diseases: microbial factors. Ann Periodontol 1996; 1: 879–925
- Hillman JD, Socransky SS, Shivers M. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch Oral Biol 1985; 30: 791–5
- van der Reijden WA, Dellemijn-Kippuw N, Stijne-van Nes AM, de Soet JJ, van Winkelhoff AJ. Mutans streptococci in subgingival plaque of treated and untreated patients with periodontitis. J Clin Periodontol 2001; 28: 686–91
- Sookkhee S, Chulasiri M, Prachyabrued W. Lactic acid bacteria from healthy oral cavity of Thai volunteers: inhibition of oral pathogens. J Appl Microbiol 2001; 90: 172–9
- Doran A, Kneist S, Verran J. Ecological control: in vitro inhibition of anaerobic bacteria by oral streptococci. Microb Ecol Health Dis 2004; 16: 23–7
- Wiebe CB, Putnins EE. The periodontal disease classification system of the American Academy of Periodontology – an update. J Can Dent Assoc 2000; 66: 594–7
- Silness J, Löe H. Periodontal disease in pregnancy. II. Correlation between oral hygiene and periodontal condition. Acta Odontol Scand 1964; 22: 121–35
- Löe H, Silness J. Periodontal disease in pregnancy. I. Prevalence and severity. Acta Odontol Scand 1963; 21: 533–51
- van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol 2002; 29: 1023–8
- WHO. Oral health surveys: basic methods4th edn. World Health Organization, Geneva 1997
- Birkhed D, Edwardsson S, Andersson H. Comparison among a dip-slide test (Dentocult®), plate count, and Snyder test for estimating number of lactobacilli in human saliva. J Dent Res 1981; 60: 1832–41
- Boström L, Linder LE, Bergström J. Influence of smoking on the outcome of periodontal surgery. A 5-year follow-up. J Clin Periodontol 1998; 25: 194–201
- Slots J. Rapid identification of important periodontal microorganisms by cultivation. Oral Microbiol Immunol 1986; 1: 48–55
- Slots J. Selective medium for isolation of Actinobacillus actinomycetemcomitans. J Clin Microbiol 1982; 15: 606–9
- Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolken RH. Manual of clinical microbiology7th edn. American Society for Microbiology, Washington, DC 1999
- Kõll-Klais P, Mändar R, Leibur E, Marcotte H, Hammarström L, Mikelsaar M. Oral lactobacilli in chronic periodontitis and periodontal health: species composition and antimicrobial activity. Oral Microbiol Immunol 2005; 20: 354–61
- Annuk H, Shchepetova J, Kullisaar T, Songisepp E, Zilmer M, Mikelsaar M. Characterization of intestinal lactobacilli as putative probiotic candidates. J Appl Microbiol 2003; 94: 403–12
- Ximénez-Fyvie LA, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol 2000; 27: 648–57
- Mikelsaar M, Mändar R, Sepp E, Annuk H. Human lactic acid microflora and its role in the welfare of the host. Lactic acid bacteria: microbiological and functional aspects, S Salminen, A von Wright, A Ouwehand. Marcel Dekker, New York 2004; 453–505
- Söder P-Ö, Jin LJ, Söder B. DNA probe detection of periodontopathogens in advanced periodontitis. Scand J Dent Res 1993; 101: 363–70
- Haffajee AD, Bogren A, Hasturk H, Feres M, Lopez NJ, Socransky SS. Subgingival microbiota of chronic periodontitis subjects from different geographic locations. J Clin Periodontol 2004; 31: 996–1002
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL, Jr. Microbial complexes in subgingival plaque. J Clin Periodontol 1998; 25: 134–44
- Griffen AL, Lyons SR, Becker MR, Moeschberger ML, Leys EJ. Porphyromonas gingivalis strain variability and periodontitis. J Clin Microbiol 1999; 37: 4028–33
- Slots J. Microflora in the healthy gingival sulcus in man. Scand J Dent Res 1977; 85: 247–54
- Dowsett SA, Kowolik MJ, Archila LA, Eckert GJ, LeBlanc DJ. Subgingival microbiota of indigenous Indians of Central America. J Clin Periodontol 2002; 29: 159–67
- Quirynen M, Gizani S, Mongardini C, Declerck D, Vinckier F, Van Steenberghe D. The effect of periodontal therapy on the number of cariogenic bacteria in different intra-oral niches. J Clin Periodontol 1999; 26: 322–7
- Tompkins GR, Tagg JR. Incidence and characterization of anti-microbial effects produced by Actinomyces viscosus and Actinomyces naeslundii. J Dent Res 1986; 65: 109–12
- Takahashi N, Schachtele CF. Effect of pH on the growth and proteolytic activity of Porphyromonas gingivalis and Bacteroides intermedius. J Dent Res 1990; 69: 1266–9