Abstract
TIP60 can act as a transcriptional activator or a repressor depending on the cellular context. However, little is known about the role of the chromodomain in the functional regulation of TIP60. In this study, we found that TIP60 interacted with H3K4me3 in response to TNF-α signaling. TIP60 bound to H3K4me3 at the promoters of the NF-κB target genes IL6 and IL8. Unlike the wild-type protein, a TIP60 chromodomain mutant did not localize to chromatin regions. Because TIP60 binds to histones with specific modifications and transcriptional regulators, we used a histone peptide assay to identify histone codes recognized by TIP60. TIP60 preferentially interacted with methylated or acetylated histone H3 and H4 peptides. Phosphorylation near a lysine residue significantly reduced the affinity of TIP60 for the modified histone peptides. Our findings suggest that TIP60 acts as a functional link between the histone code and transcriptional regulators.
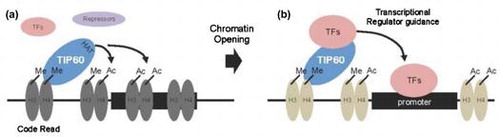
As a code translator, TIP60 interacted to methylated or acetylated histone H3, H4, inducing open chromatin state for accessibility of transcription complexes.
TIP60 (HIV Tat-interacting protein, 60 kDa; or KAT5) was identified as an interacting partner for HIV-1 Tat that increases the transcriptional activity of Tat at the HIV-1 promoter.Citation1) TIP60, which contains a MOZ, Ybf2/Sas3, SAS2, and TIP60 (MYST) domain, is a member of the MYST family of histone acetyltransferase (HAT) proteins, which are conserved from yeast to humans.Citation2) TIP60 showed intrinsic HAT activity against histone subunits H2A, H3, and H4 in vitro when purified histones were used as substrates,Citation3) which suggests that TIP60 plays roles in chromatin remodeling and gene regulation. TIP60 can function as a transcriptional coactivator or corepressor depending upon the cellular context and chromatin environment. As a coactivator, TIP60 interacts directly with numerous transcriptional activators, such as HIV-1 Tat,Citation1) type-I nuclear hormone receptors,Citation4) neural retina leucine zipper (NRL),Citation5) Pax6,Citation6) MyoD,Citation7) and NF-κB/p65.Citation8) In these instances, the coactivator function of TIP60 is mediated by its acetylation of histones at the promoter region. In other cases, TIP60 directly acetylates the transcription factor, such as p53, to modulate transcriptional activity.Citation9) On the other hand, TIP60 has also been implicated in the negative regulation of gene expression through its binding to STAT3,Citation10) cAMP response element-binding protein (CREB),Citation11) and p73βCitation12) Despite the increasing number of studies of TIP60, the roles of TIP60 in regulating gene expression from specific promoters are not well understood.
Eukaryotic DNA is packaged into chromatin, whose fundamental repeating unit is the nucleosome.Citation13) A nucleosome comprises 147 base pairs of DNA wrapped around an octamer of four core histones, H2A, H2B, H3, and H4, which are subjected to a number of post-translational modifications.Citation14) These modifications can alter chromatin structure and function by changing the charge of the nucleosome particle and/or by recruiting various protein complexes. Hence, histone modifications are thought to constitute a “histone code,” which is “read” by proteins to bring about specific downstream effects.Citation15) Individual histone modifications are associated with transcriptional activation or repression. Acetylation and phosphorylation generally accompany transcription; sumoylation and deimination are usually found in transcriptionally silent regions; and methylation and ubiquitination are implicated in the activation and repression of transcription.Citation16) Furthermore, the establishment of some modifications is dependent on the presence of other modifications. To solve the histone code, it is important to understand the specific histone modifications recognized by a code reader protein.
A growing number of proteins have been identified as transcriptional regulatory factors involved in chromatin remodeling. However, the precise mechanisms by which such regulators participate in TIP60-mediated transcriptional regulation and affect cellular physiology remain to be elucidated. In this study, we identified the histone modifications preferentially recognized by TIP60. Our results indicate that TIP60 acts as a code reader by binding to methylated histones through its chromodomain and as an effector protein through its HAT activity. TIP60 may play the role of “translator” between histone marks and downstream effectors.
Materials and methods
Cell culture and transfection
HepG2 cells were obtained from American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and penicillin–streptomycin (50 units/mL). Oligonucleotide for TIP60 siRNA was introduced into the pBabe-dual vector: forward 5′-aaag AAA CGG AAG GTG GAG GTG-3′, reverse 5′-aaaa CAC CTC CAC CTT CDCG TTT-3′. The TIP60 siRNA clone was verified by DNA sequencing. Cells were transiently transfected with different plasmid DNA constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Immunoprecipitation, western blotting, and confocal microscopy
HepG2 cells were lysed in buffer containing 1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 0.1% SDS, 1% Nonidet P-40, and 1 mM PMSF. For immunoprecipitation assays, the supernatants were precleared with 20 μL of protein A/G-agarose beads (50% slurry) and then incubated at 4 °C overnight with 30 μL of fresh protein A/G-agarose beads in the presence of the appropriate antibodies. Samples were analyzed by western blot using primary antibodies against FLAG (Sigma) and H3K4me3 (Cell Signaling Technology), followed by HRP-anti-mouse and HRP-anti-rabbit secondary antibodies (1:5000). Signal was detected by enhanced chemiluminescence (Santa Cruz Biotechnology). For confocal microscopy, HepG2 cells were washed with PBS and fixed for 1 h in 4% paraformaldehyde. The cells were permeabilized with 0.3% Triton X-100 for 20 min and incubated with 300 nM DAPI for nuclear staining. Cells were visualized using a Carl Zeiss LSM-510 META laser-scanning microscope (Oberkochen, Germany).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed following the protocol provided by Millipore (Temecula, CA, USA). Briefly, cultured HepG2 cells were cross-linked with 1% paraformaldehyde (#15710; Electron Microscopy Sciences, Hatfield, PA, USA) in PBS for 15 min at 37 °C. The cells were then washed with ice-cold PBS and resuspended in 200 μL of SDS-sample buffer containing a protease inhibitor mixture. The suspension was sonicated three times for 10 s with a 1 min cooling period on ice and then precleared for 30 min at 4 °C with 20 μL of protein A/G-agarose beads blocked with sonicated salmon sperm DNA. The beads were removed, and the chromatin solution was immunoprecipitated overnight with anti-TIP60 (sc-5725; Santa Cruz Biotechnology) and anti-histone H3K4me3 (1766-1; Epitomics, Burlingame, CA, USA) antibodies at 4 °C. The solution was then incubated with 50 μL of protein A-agarose beads (Millipore) for an additional hour at 4 °C. The immune complexes were eluted with 100 μL of elution buffer (1% SDS and 0.1 M NaHCO3), and the formaldehyde cross-links were reversed by heating at 65 °C for 4 h. Proteinase K (P2308; Sigma) was added to the reaction mixtures and incubated at 45 °C for 1 h. Immunoprecipitated DNA and control input DNA were purified using a PCR purification kit (Qiagen, Valencia, CA, USA) and then analyzed by quantitative PCR. PCR was performed in duplicate for each experimental condition tested.
Peptide array
TIP60 interacting preferences to the modified histones were tested by histone peptide array chip (Active Motif, Cat# 13001, Carlsbad, CA, USA), and procedures were followed by manufacturer’s instructions. Briefly, GST fused wild-type TIP60 was induced in E. coli after IPTG treatment and target proteins were purified using GST4B agarose bead. After blocking the peptide array chip with 5% non-fat milk in the TTBS buffer (10 mM Tris–HCl, pH 7.4, 0.05% Tween 20 and 150 mM NaCl), 500 μg of purified GST-TIP60 was incubated with peptide array chip. Protein interaction affinity was visualized by the incubation with anti-GST antibodies, followed by HRP-anti-mouse secondary antibodies (1:5000). Signal was detected by enhanced chemiluminescence (Santa Cruz Biotechnology).
Results
TIP60 localizes to the active promoter region of the IL6 and IL8 genes through interaction with H3K4me3
TIP60 plays critical roles in apoptosis, development, and damage responses.Citation17,18) However, the role of TIP60 as a chromatin-remodeling factor is not well understood. Previously, we reported that TIP60 was present at the promoter region of target genes such as IL6, IL8, Rho, and Ppp2r5c before key transcription factors, NF-κB (RelA/p65 subunit) and NRL, were recruited.Citation5,8) We proposed that TIP60 acts as a “histone code reader” that makes chromatin accessible to transcription factors. In this study, to test the hypothesis that TIP60 affected RelA/p65-dependent transcriptional activity through interaction with H3K4me3, we expressed FLAG-TIP60 in HepG2 cells and treated the cells with TNF-α. Whole cell lysates were immunoprecipitated with an anti-H3K4me3 antibody, and endogenous H3K4me3 and overexpressed TIP60 levels were assessed by western blot with specific anti-FLAG and anti-H3K4me3 antibodies. FLAG-TIP60 coimmunoprecipitated with H3K4me3 (Fig. (A); lane 2), but did not coimmunoprecipitate with control FLAG-empty vector (Fig. (A), lane 1). The interaction between FLAG-TIP60 and H3K4me3 increased about twofold when cells were treated with TNF-α (Fig. (A), lane 3).
Fig. 1. TIP60 associates with the IL6 and IL8 gene promoters through direct interaction with H3K4me3.
Notes: (A) HepG2 human hepatocarcinoma cells were transfected with plasmids expressing FLAG-tagged TIP60 (1 μg). After 48 h of transfection, HepG2 cells were treated with 50 ng/mL TNF-α for 30 min. Whole cell lysates were then immunoprecipitated with an anti-H3K4me3 antibody, and the precipitated fractions were analyzed by western blot using antibodies against FLAG and H3K4me3. The band intensity was measured and precipitated TIP60 levels were normalized by precipitated H3K4me3. (B) Schematic diagram of primer pairs in the distal and proximal region, and κB-element containing regions of the IL-6 and IL-8 promoter used in ChIP assay. (C) HepG2 cells were transfected with TIP60 siRNA (white box) or empty vector (black box). Forty-eight hours after transfection, HepG2 cells were treated with 50 ng/mL TNF-α for the indicated times (0, 15, 30, 45, or 60 min) and processed for ChIP assays with anti-TIP60 and anti-H3K4me3 antibodies. Immunoprecipitated chromatin was analyzed by quantitative PCR using primers specific for the IL6 and IL8 promoters. All values are shown as the mean ± SE of three independent experiments.
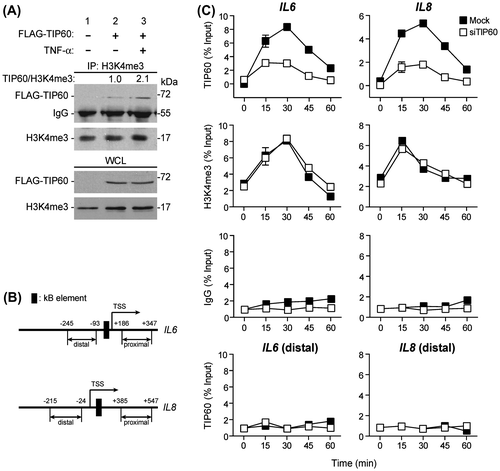
Having established that TIP60 interacted with histone H3K4me3, we used ChIP assays to examine whether TIP60 was recruited to the promoters of NF-κB target genes in vivo. We stimulated HepG2 cells with TNF-α for 0, 15, 30, 45, or 60 min and examined the binding of TIP60 and H3K4me3 to the IL6 and IL8 promoters. In the absence of TNF-α stimulation (0 min in each experimental group), the binding of TIP60 and H3K4me3 to each promoter was barely detectable. After TNF-α stimulation (0–30 min), H3K4me3 and TIP60 were recruited to the promoters of the IL6 and IL8 genes (Fig. (C)) whereas distal and proximal regions of these promoters which have no NF-κB-binding sequences were not specifically precipitated by TIP60 antibody (Fig. (B), and (C) and data not shown). Interestingly, the pattern of H3K4me3 binding to IL6 and IL8 promoters was not affected by the presence of TIP60 (Fig. (C) and Supplemental Fig. 2(A)). H3K4me3 binding did not change when cells were treated with TIP60 siRNA. These results suggest that histone modification induced by NF-κB signaling occurs in a TIP60-independent manner. TIP60 then binds to the target gene promoters through recognition of histone H3 Lys4 methylation.
Mutation of the TIP60 chromodomain causes mislocalization
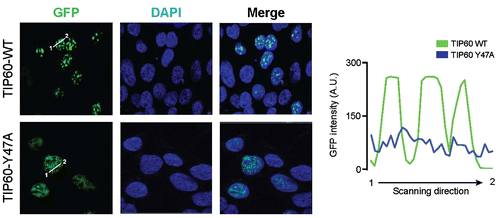
To examine the function of the TIP60 chromodomain, we transfected cells with plasmids encoding wild-type GFP-TIP60 (TIP60-WT) or a chromodomain mutant (GFP-TIP60-Y47A). The localization of the GFP-tagged TIP60 proteins was visualized by confocal microscopy. TIP60-WT localized to the chromatin-rich region in a few foci with highly concentrated intensity in the nucleus (Fig. ). However, the GFP-TIP60-Y47A mutant was spread throughout the nucleus, over a hundred small punctae were visible with weak intensity compared to TIP60-WT. These results suggest that TIP60 interacts with chromatin and that the chromodomain of TIP60 mediates chromatin recognition.
Fig. 2. Mutation of the TIP60 chromodomain causes mislocalization.
Notes: HepG2 cells were transfected with plasmids expressing wild-type GFP-TIP60 (TIP60-WT) or a GFP-TIP60 chromodomain mutant (TIP60-Y47A). GFP fluorescence was examined by confocal microscopy and fluorescence signal intensity was quantified using line scanning method. DAPI was used to counterstain nuclei.
Screening for interactions between TIP60 and specific histone modifications
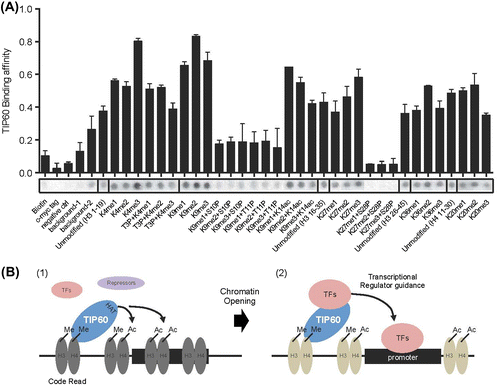
To assess TIP60 binding to different histone modifications, we used a histone peptide chip that included 379 modified histone peptides and 5 control peptides. GST-TIP60 was expressed in bacteria, purified, and incubated with peptide the chip for 2 h. The interaction of GST-TIP60 with the histone peptides was detected using GST antibodies. The intensity of the interactions was measured by analyzing scanned images with Array Analyzer Software version 15 (Supplementary data 1). Two replicates are shown in Fig. , their similarity is evident. Compared to the negative controls, the interaction value of binding peptides were determined over 0.374 (below 0.374 is background) (Fig. (B)). The interaction values for the 167 histone peptides that interacted with TIP60 ranged from 0.37 (H3K27me2, H3K28p) to 1 (H4K12, K16, and K20 tri-acetylated peptides). TIP60 interacted with 46 and 52% of the modified H3 and H4 histone peptides included in the array (total H3 peptides: 273; total H4 peptides: 69). In contrast, TIP60 interacted with 27 and 13% of the modified H2A and H2B peptides (total H2A peptides: 15; total H2B peptides: 15) (Fig. (C)). To determine whether TIP60 preferentially interacted with specific histone modifications, we examined the strongest and the weakest interacting peptides. As shown in Fig. (D), TIP60 preferentially interacted with histone H3 and H4 peptides that were acetylated or methylated. However, phosphorylation significantly inhibited the interaction of TIP60 with histone H3 and H4 peptides, even when the peptides were acetylated or methylated (Figs. (D) and (A)). These results suggest that TIP60 preferentially interacts with acetylated or methylated histones H3 and H4, rather than with histones H2A and H2B, and that TIP60 binds weakly to phosphorylated histones.
Fig. 3. TIP60 preferentially binds acetylated or methylated histone H3 and H4 peptides.
Notes: (A) Two different data-sets were compared. R-squared value indicates 0.91 between the two biological replicates. (B) The affinity of TIP60 for differentially modified histone peptides was visualized by arranging the peptides in order from strongly interacting to weakly interacting (1–0) using Array Analyze Software version 15. Of 384 different peptides, 167 peptides generated an interaction signal above the background level (>0.374). (C) The percentage of interacting peptides among all histone H3, H4, H2A, and H2B peptides is shown. TIP60 bound preferentially to specific histone modifications. (D) The top 10 strongly interacting peptides and weakly interacting peptides are shown. All values are presented in Supplementary data 1.
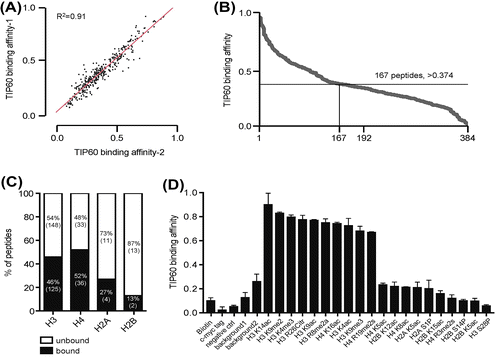
Fig. 4. Phosphorylation of histone H3 on Ser10, Thr11, and Ser28 inhibits the interaction between TIP60 and acetylated or methylated histone H3 peptides.
Notes: (A) The peptide array shows the affinity of TIP60 for specific combinations of histone modifications. Each spots represent the affinity between TIP60 and a histone peptide. (B) Schematic model of transcriptional regulation by TIP60. TIP60 is recruited to a methylated histone region through its chromodomain. TIP60 then acetylates nearby lysine residues to open the compact chromatin (1). Depending on the cellular context and the chromatin environment, TIP60 recruits transcriptional activators or repressors via protein–protein interactions to modulate target gene expression. Acting as a mediator between the histone code and TFs, TIP60 enhances the loading of TFs onto the promoter region (2).
Binding of TIP60 to histone H3 and H4 peptides with different combinations of modifications
Because the binding of TIP60 to acetylated or methylated histone H3 and H4 peptides was inhibited by phosphorylation, we investigated the combinatorial effects of histone H3 and H4 acetylation, methylation, and phosphorylation on TIP60 binding. As shown in Fig. , the TIP60 binding affinity for H3K4me3 (binding value: 0.80) was stronger than the binding affinity for H3K4me1/2 (binding value for me1: 0.56, for me2: 0.53) or the unmodified H3 peptide (binding value: 0.37). However, phosphorylation on Thr3 (T3P) reduced binding affinity of TIP60 for H3K4me3 (Fig. ). TIP60 interacted with mono-, di-, and trimethylated H3K9. However, phosphorylation on Ser10 and Ser11 markedly reduced the interaction between TIP60 and the H3K9 methylated peptides. Acetylation on Lys14 did not markedly affect H3K9me modification. As with H3K4 and H3K9, H3K27 methylation levels were affected by phosphorylation on Ser28. The TIP60 binding affinities for H3K36me2 and H4K20me2 were slightly higher than the affinities for the unmodified control peptides. These results show that the phosphorylation status of Ser/Thr residues near Lys4, Lys9, and Lys27 affects the interaction between TIP60 and methylated histone H3 peptides.
Discussion
Growing evidence suggests that TIP60 plays an important role in the regulation of transcription factor activity by acting as coactivator or corepressor. However, the mechanism by which TIP60 contributes to the regulation of gene expression is not well characterized. TIP60 does not recognize specific DNA sequences directly. Rather, it requires the selective activity of transcription factors or regulatory proteins to modulate the expression of specific target genes. In this study, TIP60 interacted with histone H3K4me3 and regulated the expression the NF-κB target genes IL6 and IL8 through its association with the target-gene promoter region (Fig. ). The cellular distribution of TIP60 was consistent with recruitment to specific chromatin regions. However, the cellular distribution of a TIP60 chromodomain mutant differed from that of the wild-type protein, with a scattered localization throughout the nucleus (Fig. ). This suggests that TIP60 associates with chromatin through its chromodomain to regulate target gene expression. To assess the functional importance of TIP60 in chromatin remodeling, we first designed a TIP60 knock-in (KI) system using transcription activator-like effector nucleases for genome engineering. However, TIP60-KI cells did not divide properly after selection, and the cells died after 3–4 cell divisions (Supplementary data 2(B)–(D)). This result is consistent with the finding that a Tip60 knockout mouse line.Citation19)
Chromodomains are specialized binding modules that contain conserved hydrophobic amino acids, which interact with the methyl groups on methylated lysine residues.Citation20,21) For example, the chromodomain in HP1α interacts with H3K9me3.Citation20) Sequence alignment of the chromodomains of TIP60 and HP1α showed that TIP60 contains the conserved aromatic amino acids that are required for the binding of HP1α to H3K9me3, implying that the chromodomain of TIP60 functions as a methyl-lysine binding domain.Citation22) TIP60 also has intrinsic HAT activity. Previous studies demonstrated that TIP60 interacts with transcription factors or transcriptional regulators to modulate target gene expression.Citation17,18) We propose that TIP60 functions as a “code translator” between histone modifications and TIP60-interacting transcriptional regulators (Fig. (B)). Several findings in this study support this proposal. First, TIP60 recognized specialized histone codes on chromatin and localized to appropriate regions through its chromodomain (Fig. (B); (1)). Second, TIP60 opened closed, compact chromatin by directly acetylating histone proteins (Fig. (B); (1)). Third, TIP60 recruited other transcriptional regulators to modulate target gene expression (Fig. (B); (2)). The effect on gene expression would depend on whether the recruited proteins were activators or repressors, which is determined by the cellular context and chromatin niche.
To identify the histone codes for TIP60, we used a histone peptide array chip containing histone peptides with different modifications (Fig. ). As shown in previous reports, TIP60 interacted preferentially with histones H3 and H4 and more strongly with acetylated or methylated histones.Citation3) Interestingly, histone phosphorylation near TIP60 binding sites significantly inhibited the interaction of TIP60 with the histone peptides (Fig. ). Histone phosphorylation is involved in transcriptional regulation, DNA damage repair responses, and chromatin compaction during cell division.Citation23,24) These processes often involve the regulation of proliferative genes. Phosphorylation of Ser10 and 28 in H3 and Ser32 in H2B is associated with the regulation of epidermal growth factor-responsive gene transcription. H3S10ph and H2BS32ph are linked to the expression of proto-oncogenes such as c-Fos and c-Myc. Phosphorylation of H3S10, T11, and S28 are clearly associated with histone H3 acetylation, which implicates these modifications in transcription activation. Independent studies have shown that these phosphorylation events are mechanistically linked to Gcn5-dependent histone H3 acetylation. However, TIP60 binding and histone phosphorylation were mutually exclusive in our studies (Fig. (A)). It is possible that once histones are acetylated by HAT, subsequent phosphorylation inhibits the recruitment of other HATs to the specific promoter region. Further molecular studies are required to define the mechanisms that connect histone acetylation and phosphorylation.
Chromatin remodeling through various histone modifications is mediated by protein–protein interactions between histones and transcription factors and by protein–DNA interactions. This study adds to our understanding of this process by elucidating the role of TIP60 in transcriptional regulation. Furthermore, regulating the interactions between TIP60 and histone modifications might be a way to target the systemic activation or inhibition of the TIP60 pathway.
Supplemental material
The supplemental material for this paper is available at http://dx.doi.org/10.1080/09168451.2014.993914.
Supplemental Materials
Download Zip (196.6 KB)Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) [grant number 2012008662].
Additional information
Funding
References
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366.10.1006/viro.1996.0071
- Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976.10.1093/nar/gkh252
- Kimura A, Horikoshi M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells. 1998;3:789–800.10.1046/j.1365-2443.1998.00229.x
- Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, Robson CN. Tip60 is a nuclear hormone receptor coactivator. J. Biol. Chem. 1999;274:17599–17604.10.1074/jbc.274.25.17599
- Kim JW, Jang SM, Kim CH, An JH, Choi KH. Transcriptional activity of neural retina leucine zipper (Nrl) is regulated by c-Jun N-terminal kinase and Tip60 during retina development. Mol. Cell Biol. 2012;32:1720–1732.10.1128/MCB.06440-11
- Kim CH, Kim JW, Jang SM, An JH, Song KH, Choi KH. Transcriptional activity of paired homeobox Pax6 is enhanced by histone acetyltransferase Tip60 during mouse retina development. Biochem. Biophys. Res. Commun. 2012;424:427–432.10.1016/j.bbrc.2012.06.126
- Kim JW, Jang SM, Kim CH, An JH, Kang EJ, Choi KH. Tip60 regulates myoblast differentiation by enhancing the transcriptional activity of MyoD via their physical interactions. FEBS J. 2011;278:4394–4404.10.1111/j.1742-4658.2011.08362.x
- Kim JW, Jang SM, Kim CH, An JH, Kang EJ, Choi KH. New molecular bridge between RelA/p65 and NF-kappaB target genes via histone acetyltransferase TIP60 cofactor. J. Biol. Chem. 2012;287:7780–7791.10.1074/jbc.M111.278465
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 2006;24:841–851.10.1016/j.molcel.2006.11.026
- Xiao H, Chung J, Kao HY, Yang YC. Tip60 is a co-repressor for STAT3. J. Biol. Chem. 2003;278:11197–11204.10.1074/jbc.M210816200
- Gavaravarapu S, Kamine J. Tip60 inhibits activation of CREB protein by protein kinase A. Biochem. Biophys. Res. Commun. 2000;269:758–766.10.1006/bbrc.2000.2358
- Kim JW, Song PI, Jeong MH, An JH, Lee SY, Jang SM, Song KH, Armstrong CA, Choi KH. TIP60 represses transcriptional activity of p73beta via an MDM2-bridged ternary complex. J. Biol. Chem. 2008;283:20077–20086.10.1074/jbc.M800161200
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871.10.1126/science.184.4139.868
- Thomas JO, Kornberg RD. An octamer of histones in chromatin and free in solution. Proc. Nat. Acad. Sci. USA. 1975;72:2626–2630.10.1073/pnas.72.7.2626
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080.10.1126/science.1063127
- Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 20:259–266.
- Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 2006;38:1496–1509.10.1016/j.biocel.2006.03.003
- Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9:930–936.10.4161/cc.9.5.10931
- Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev. Dyn. 2009;238:2912–2921.10.1002/dvdy.v238:11
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107.10.1038/nature722
- Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, Grant PA 3rd. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438.10.1038/nature03242
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 2009;11:1376–1382.10.1038/ncb1982
- Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220.10.1016/j.tig.2004.02.007
- Rossetto D, Avvakumov N, Cote J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics. 7:1098–1108.
