Abstract
Speradine A is a derivative of cyclopiazonic acid (CPA) found in culture of an Aspergillus tamarii isolate. Heterologous expression of a predicted methyltransferase gene, cpaM, in the cpa biosynthesis gene cluster of A. tamarii resulted in the speradine A production in a 2-oxoCPA producing A. oryzae strain, indicating cpaM is involved in the speradine A biosynthesis.
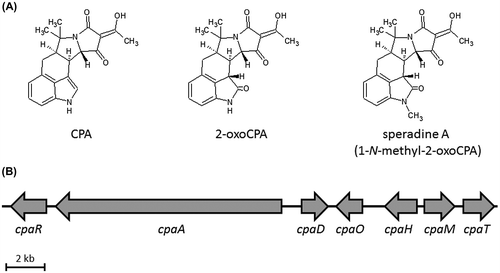
Cyclopiazonic acid (CPA) (Fig. (A)) is an indole tetramic acid that functions as an inhibitor of sarcoplasmic or endoplasmic reticulum calcium-dependent ATPase (SERCA).Citation1,2) Some strains in the genera Aspergillus and Penicillium have been reported to produce CPA,Citation3) and the CPA derivatives 2-oxoCPA and speradine A (1-N-methyl-2-oxoCPA) (Fig. (A)) are produced by Aspergillus oryzae NBRC 4177Citation4) and A. tamarii marine-derived isolate,Citation5) respectively. The CPA biosynthesis gene cluster (cpa cluster; Fig. (B)) in A. oryzae is composed of 7 genes: cpaR, cpaA, cpaD, cpaO, cpaH, cpaM, and cpaT.Citation3) The cpaA, cpaD, and cpaO genes are required for CPA biosynthesis.Citation6–8) We previously demonstrated that cpaH is involved in the modification of CPA to 2-oxoCPA.Citation4) The other genes, cpaR, cpaM, and cpaT, are predicted to encode a C6-type transcription factor, methyltransferase, and major facilitator superfamily (MFS) transporter protein,Citation4) respectively, but have not yet been characterized because gene disruption did not affect CPA production in A. oryzae.Citation4) There is a possibility that cpaM is originally involved in the biosynthesis of speradine A since it could be biosynthesized via methylation of 2-oxoCPA. Thus, we inferred that the inability of speradine A production in A. oryzae is caused by mutations or partial deletions in cpaM. In this study, to clarify the function of cpaM, we analyzed speradine A production in an A. oryzae strain expressing a cpaM homolog from speradine A-producing A. tamarii strain.
Fig. 1. Chemical structure of CPA,Citation1) 2-oxoCPACitation4) and speradine A (1-N-methyl-2-oxoCPA)Citation5) (A) and CPA biosynthesis gene cluster (cpa cluster) in Aspergillus oryzae NBRC 4177 (B).Citation4)
We obtained the CPA-producing A. tamarii strain NBRC 4099 from the Biological Resource Center, NITE (Kisarazu, Japan), and verified its speradine A production. Strain NBRC 4099 was cultured statically on CYA liquid medium (0.3% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO47H2O, 0.001%FeSO47H2O, 0.5% yeast extract, and 3.0% sucrose) for 6 days, and the metabolites in the culture were extracted with an equal volume of ethyl acetate. The extracted metabolite sample and authentic speradine ACitation5) diluted in ethyl acetate to 10 ppm were analyzed by high-performance liquid chromatography–electrospray ionization (+)-time-of-flight mass spectrometry (1260 Infinity and G6500; Agilent Technologies, Santa Clara, CA, USA). For the mobile phases, 0.1% (v/v) formic acid water (eluent A) and acetonitrile (eluent B) were used. Metabolites (3 μl) extracted from fungal cultures were separated with a C18 column (L-column ODS,2.1 × 100 mm, 5 μm particle size, Chemical Evaluation and Research Institute, Tokyo, Japan) with gradient elution (0–5 min, 0% B; 5–20 min, 0–100% B; 20–25 min, 100% B; 25–26 min, 100–0% B; 26–36 min, 0% B). The extracted ion chromatograms of m/z 367.165 (corresponding to the [M+H]+ values of speradine A) showed distinct peaks at 17.3 min in both the sample of the metabolite sample of A. tamarii NBRC 4099 and authentic speradine A (Supplemental Fig. 1).
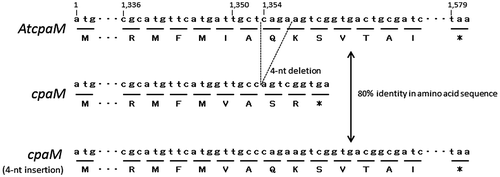
Next, we partially sequenced the cpa cluster in A. tamarii because the genomic sequence of A. tamarii was not available. Using PCR primers based on cpaH, cpaM, cpaT, and their flanking region in A. oryzae, we obtained amplified products from the genomic DNA of A. tamarii NBRC 4099. We successfully sequenced 6,394 bp of the genomic region and found cpaH, cpaM, and cpaT homologs in A. tamarii (referred to as AtcpaH, AtcpaM, and AtcpaT, hereafter). The nucleotide identity between the sequenced region in A. tamarii and the corresponding region in A. oryzae was 78.9%. The coding region between AtcpaH and cpaH, AtcpaM and cpaM, and AtcpaH and cpaH showed 86, 83, and 88% nucleotide sequence identities and the query coverage is 100, 99, and 99%, respectively. We previously suggested that the genomic region spanning cpaH and cpaM was unique to A. oryzae,Citation3) but A. tamarii NBRC 4099 also possessed this genomic region.
Since an A. tamarii transformation system has not been established, we were unable to construct a gene-knockout strain of AtcpaM. Instead, we introduced AtcpaM into an A. oryzae strain to clarify the gene’s involvement in speradine A biosynthesis. A transformation vector carrying AtcpaM and its flanking regions was constructed using the binary vector pPTRI (Takara, Shiga, Japan), which harbors a fungal pyrithiamine-resistant gene, ptrA.Citation9) The 2482-bp genomic region containing the predicted coding sequence of AtcpaM, 5′-intergenic region (376 bp) of the predicted AtcpaM and adjacent AtcpaH, and 3′-flanking region (524 bp), was amplified by PCR (KOD plus Neo, TOYOBO, Osaka, Japan) according to the manufacturer’s instructions with the following primers: 5′-CCAAGCTTGCATGCCTTTGGGTAGATTCAGGCGAACAGGTAGAC-3′, 5′-CCTCTAGAGTCGACCGGCGGCTCTTCCTGGTGACCCATG -3′. The pPTRI fragment was amplified by PCR with the primers 5′-GGTCGACTCTAGAGGATCCCCGGGTACCG-3′ and 5′- GGCATGCAAGCTTGGCGTAATCATGGTC-3′. The amplified DNA fragments of the AtcpaM with flanking region and the pPTRI were combined using the In-Fusion system (Takara), according to the manufacturer’s instruction. The resulting transformation vector pPAtcM was introduced into a 2-oxoCPA-producing A. oryzae strain, NBRC 4177Citation4) by the protoplast transformation method.Citation10) Regenerated fungal isolates grown in selective medium were checked for integration of the expression vector in the genomic DNA by PCR analysis using the primer set for AtcpaM and its flanking region amplification. As shown in Fig. (A), amplified fragments that coincided with the size of AtcpaM (2,482 bp) were detected in the lanes of transformants PAtcM-5, and -6 and A. tamarii NBRC 4099, but no fragment was observed in the lane of the recipient strain, A. oryzae NBRC 4177, confirming that transformants PAtcM-5, and -6 harbored the AtcpaM and its flanking regions.
Fig. 2. Production of the CPA and the other derivatives in the transformants.
Notes: (A) Confirmation of transformation by PCR amplification of the introduced AtcpaM gene in the constructed strains, PAtcM-5 and -6. (B) UV chromatogram of 280 nm for the metabolite samples extracted from the culture broth of the transformants (PAtcM-5 and -6), A. oryzae NBRC 4177, and A. tamarii NBRC 4099. (C) The extracted ion chromatograms of m/z 337.155 (CPA), m/z 353.150 (2-oxoCPA), and m/z 367.165 (speradine A) in the metabolite samples of the transformants (PAtcM-5 and -6), A. oryzae NBRC 4177, and A. tamarii NBRC 4099. (D) MS/MS fragmentation analysis of the ions with of m/z 367.165 detected in the sample of A. oryzae transformant PAtcM-6, A. tamarii NBRC 4099 and the authentic speradine A. The collision energy was set to 24.5 eV.
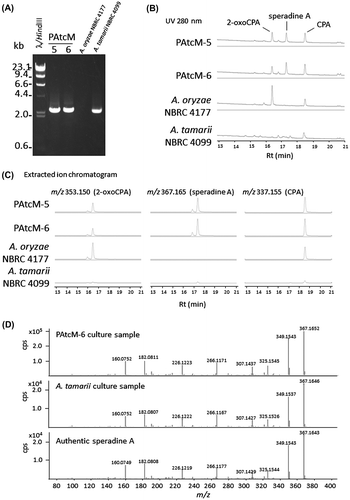
Two transformants (PAtcM-5 and -6), the recipient strain (A. oryzae NBRC 4177), and A. tamarii NBRC 4099 were cultured statically in CYA liquid medium for 6 days and metabolites extracted by ethyl acetate were analyzed by high-performance liquid chromatography-electrospray ionization (+)-time-of-flight mass spectrometry, as mentioned above. The UV chromatogram and the extracted mass chromatogram of m/z 337.155 and 353.150 corresponding to [M+H]+ values of CPA and 2-oxoCPA, respectively, were clearly detected at Rt = 16.3 min and Rt = 18.5 min in the sample of A. oryzae NBRC 4177 (Fig. (B) and (C)), as reported previously.Citation4) In addition to CPA and 2-oxoCPA, chromatogram of the samples of two A. oryzae transformant harboring AtcpaM (PAtcM-5 and -6) showed distinct peak at Rt = 17.3 min (Fig. (B) and (C)), which is corresponding to speradine A (Supplemental Fig. 1). Further analysis by MS/MS fragmentation of the ions with m/z 367.165 eluted at 17.3 min in the sample of PAtcM-6, A. tamarii NBRC 4099 and authentic speradine A showed an identical fragmentation pattern, confirming that the compound produced by PAtcM-6 and A. tamarii NBRC 4099 were speradine A (Fig. (D)). Thus, it is indicated that AtcpaM encodes an enzyme required for speradine A synthesis. The speradine A production levels of PAtcM-5 and 6 were higher than that of A. tamarii strain (Fig. (B) and (C)), which is probably due to the lower CPA and 2-oxoCPA production level of A. tamarii strain in this culture condition. We also found several UV absorbing metabolites in the culture of A. tamarii NBRC 4099, even though we could not identified any of them in this study. In the extracted ion mass chromatograms, additional peaks eluted 0.5 min prior to 2-oxoCPA and speradine A were detected in the sample of A. oryzae strains (Fig. (C)). This suggests the presence of a structural isomer of 2-oxoCPA and its methyl derivative synthesized by enzyme encoded by cpaM, although more detailed analysis is required.
In addition, we did not detect signals corresponding to 1-N-methyl-CPA, which is deduced to be synthesized by 1-N-methylation of CPA. Thus, speradine A is suggested to be synthesized via 2-oxoCPA. Recently, new CPA-like metabolites, namely speradine F, G, and H, were found in the culture broth of marine-derived A. oryzae isolates.Citation11) However, peaks with m/z 415.151, 287.139, or 373.116, corresponding to speradine F, G, and H, were not found for the extract of the culture broth of either the A. tamarii or A. oryzae strains. The culture conditions may be critical for the production of these derivatives, or nonenzymatic generation during metabolite extraction might occur.
A. oryzae NBRC 4177 does not produce speradine A, suggesting cpaM does not function. However, a comparison of the genomic DNA sequences of cpaM and AtcpaM did not provide clear evidence for the lack of cpaM function. To identify the underlying cause of the nonfunctionality of cpaM in A. oryzae, we compared the cDNA sequences of cpaM and AtcpaM. Both A. oryzae NBRC 4177 and A. tamarii NBRC 4099 were cultured on CYA medium for 2 days and total RNAs were extracted with Isogen (Nippon Gene, Toyama, Japan) according to the manufacturer’s instructions. After DNase I (Toyobo) treatment of the extracted total RNA (10 μg), cpaM and AtcpaM cDNA were synthesized by reverse transcription with Superscript II (Invitrogen, Waltham, MA, USA) using oligo dT primers. The nucleotide sequences of the cDNAs of cpaM and AtcpaM showed that AtcpaM encodes a protein with 462 amino acids while cpaM encodes a protein with 389 amino acids. The amino acid sequences of the cpaM and AtcpaM showed 84% coverage and 78% identities. Interestingly, the difference is mainly attributed to a 4-nucleotide (CAGA) deletion in cpaM at the position that is 1,354 bp downstream of the start codon of AtcpaM (Fig. ). The deletion results in a frameshift and creates stop codon 6 nucleotides downstream. When the position of the deletion included the 4 nucleotides (i.e. when the deletion was absent), cpaM encoded a protein with 461 amino acids, which had 100% coverage and 80% identity to AtcpaM at the amino acid level (Fig. ). Thus, the 4-nucleotide deletion in cpaM likely explains the inactive cpaM. However, methyltransferase_18 domain (Pfam12847) containing S-adenosylmethionine binding site was predicted as the only functional motif by NCBI’s conserved domain database both in the proteins encoded by cpaM and AtcpaM, indicating that 4-nucleotide deletion does not lead to loss of the functional domain in cpaM protein. Further analysis to identify the determinant of inactive cpaM and enzymatic characterization using the purified protein is required; our data strongly suggest that cpaM in the cpa cluster encodes an N-methyltransferase involved in speradine A synthesis.
Authors contribution
M.T. designed the study and wrote the manuscript; T.K., Y.S., A.K., and S.I. performed experiments; T.K. and J.K. purified and provided speradine A; Y.K., A.T., H.S., and K.S. gave conceptual advice.
Supplementary material
The supplementary material for this article is available online at http://dx.doi.org/10.1080/09168451.2015.1065167.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental_Fig_caption.docx
Download MS Word (14.6 KB)SupplementalFig1.TIF
Download TIFF Image (27.8 KB)Notes
Abbreviations: CPA - cyclopiazonic acid; 2-oxoCPA, 2-oxocyclopiazonic acid
References
- Hopzfel CW. The isolation and structure of cyclopiazonic acid, a toxic metabolite of Penicillium cyclopium westling. Tetrahedron. 1967;24:2101–2119.
- Goeger DE, Riley RT, Dorner JW, Cole RJ. Cyclopiazonic acid inhibition of the Ca2+-transport ATPase in rat skeletal muscle sarcoplasmic reticulum vesicles. Biochem. Pharmacol. 1988;37:978–981. 10.1016/0006-2952(88)90195-5
- Chang PK, Ehrlich K, Fujii I. Cyclopiazonic acid biosynthesis of Aspergillus flavus and Aspergillus oryzae. Toxins. 2009;1:74–99.10.3390/toxins1020074
- Kato N, Tokuoka M, Shinohara Y, et al. Genetic safeguard against mycotoxin cyclopiazonic acid production in Aspergillus oryzae. ChemBioChem. 2011;12:1376–1382.10.1002/cbic.v12.9
- Tsuda M, Mugishima T, Komatsu K, et.al. Speradine A, a new pentacyclic oxindole alkaloid from a marine-derived fungus Aspergillus tamarii. Tetrahedron. 2003;59:3227–3230.10.1016/S0040-4020(03)00413-7
- Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y. Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet. Biol. 2008;45:1608–1615.10.1016/j.fgb.2008.09.006
- Seshime Y, Juvvadi PR, Tokuoka M, et al. Functional expression of the Aspergillus flavus PKS-NRPS hybrid CpaA involved in the biosynthesis of cyclopiazonic acid. Bioorg. Med. Chem. Lett. 2009;19:3288–3292.10.1016/j.bmcl.2009.04.073
- Chang PK, Horn BW, Dorner JW. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet. Biol. 2009;46:176–182.10.1016/j.fgb.2008.11.002
- Kubodera T, Yamashita N, Nishimura A. Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 2000;64:1416–1421.10.1271/bbb.64.1416
- Gomi K, Iimura Y, Hara S. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric. Biol. Chem. 1987;51:2549–2555.10.1271/bbb1961.51.2549
- Hu X, Xia QW, Zhao YY, et al. Speradines F–H, three new oxindole alkaloids from the marine-derived fungus Aspergillus oryzae. Chem. Pharm. Bull. 2014;62:942–946.10.1248/cpb.c14-00312