Abstract
12-Oxo-phytodienoic acid (OPDA) is induced by mechanical wounding and suppresses the growth of Physcomitrella patens; OPDA is considered as a signal compound in this moss species. In this study, a proteomic analysis of P. patens protonemata treated with OPDA was performed. The abundance levels of 41 proteins were significantly altered by OPDA, with decreased levels for 40 proteins. The proteins for which abundance decreased in response to OPDA at the protonema developmental stage were mainly involved in the metabolism of proteins and carbohydrates. The effects of inhibition on protein abundance are likely a major physiological function of OPDA in P. patens. OPDA also suppressed the expression of histones at the protein level and gene transcription level. Suppression of histone expression might be an OPDA-specific function in P. patens protonemata. In P. patens, a subset of the physiological responses caused by OPDA is shown to differ between protonema and gametophore developmental stages.
Graphical abstract
Classification of OPDA-regulated proteins in Physcomitrella patens protonema according to their functions.
The jasmonates (i.e. jasmonic acid (JA)-related compounds) are synthesized from the fatty acid of the octadecanoid pathway. Jasmonates modulate the expression of numerous genes and mediate responses to various forms of abiotic and biotic stresses.Citation1–4) 12-Oxo-phytodienoic acid (OPDA) is a precursor of JA, a plant growth regulator, and plays pivotal roles in plants.Citation5) All of the genes encoding JA biosynthetic enzymes have been cloned from several plant species. The corresponding enzymes, including different isoforms, have been characterized.Citation6) Previous reports have described the cloning of genes encoding LOX, AOS, and AOC in the octadecanoid pathway of P. patens; the products of these genes have been shown to have enzymatic activities similar to those in flowering plants.Citation7–10) In flowering plants, JA stimulates the expression of a wide variety of genes in response to stress.Citation11) OPDA itself triggers the expression of a set of genes and plays roles in the response to wounding in Arabidopsis.Citation12) Moreover, OPDA has been reported to play an important role in embryo development and seed germination.Citation13,14)
Bryophytes, which include liverworts, hornworts, and mosses, are considered to be some of the first land plants. It is important to elucidate the physiology of bryophytes to understand plant evolutionary processes. Among the bryophytes, P. patens is the model moss species. Protein and gene databases and facile gene transformation methods have already been established for P. patens.Citation15) The life cycle of P. patens is characterized by the following two generations: a haploid gametophyte and diploid sporophyte. A spore develops into a filamentous structure known as protonema. Protonemata can differentiate into gametophores; gametophores are more complex, containing leaf-like structures, rhizoids, and sexual organs. As protonemata are distinct from gametophores, gene expression is altered. Therefore, the protein abundance profile in the protonema stage is considered different from that in the gametophore stage.Citation16)
Plant hormones regulate a wide variety of physiological events in plants. Studies have demonstrated that plant hormones, such as auxin, cytokinins, and abscisic acid, also control physiological responses in P. patens. In P. patens, the first half of the octadecanoid pathway exists; however, JA is not synthesized.Citation10) OPDA inhibits the growth of P. patens.Citation17) Moreover, OPDA was induced when wounding occurs in P. patens.Citation18) Thus, OPDA may be an important oxylipin and act as a signaling molecule in P. patens.
A proteomic analysis of P. patens gametophores treated with OPDA was conducted to investigate the function of OPDA.Citation19) A previous report revealed that OPDA treatment of gametophores resulted in the differential accumulation of several proteins, most of which decreased. As the expression of genes and proteins is altered when differentiation from protonema to gametophore occurs,Citation16) we are interested in whether the effects of OPDA on protein abundance are different at these two stages of development.
In this study, a proteomic analysis of P. patens protonemata treated with OPDA was conducted. These data were compared with proteomic data from P. patens gametophores. Our findings suggest that OPDA mainly inhibits protein metabolism, which might retard the growth of P. patens.
Materials and methods
Plant growth conditions and treatment
The wild-type strain of P. patens subsp. patens was used in this study.Citation20) P. patens was grown on 20 mL of BCDATG agar medium in a 9-cm Petri dish under continuous white fluorescent light at 25 °C.Citation19) For the microscopic analysis of protonema growth, protonemata grown on the agar plate for four days were cut into small pieces using a homogenizer (Polytoron PT-10, Kinematica AG, Luzern, Switzerland). After being transferred onto agar supplemented with OPDA at various concentrations, protonemata were incubated for four days, after which the phenotypes were observed with a microscope (BZ-9000 fluorescence microscope, Keyence, Osaka, Japan). (+)-cis-OPDA synthesis was conducted according to the method described by Kajiwara et al.Citation21)
Analysis of OPDA concentration
P. patens was grown on BCDATG agar medium for five days, and the concentration of endogenous OPDA was analyzed. Protonemata (approximately 200 mg) were frozen in liquid nitrogen and extracted with 10 mL of ethanol. The OPDA analysis was performed as described by Yamamoto et al.Citation22) For mechanical stress, the protonemata (approximately 200 mg) were soaked in 2 mL of water and subsequently treated with ultrasonication (Sonifier250, Branson, USA). The samples were directly extracted with 10 mL of ethanol.
Protein extraction
P. patens protonemata were grown on BCDATG agar plates for five days and then treated with 10 μM OPDA (1 mL of an OPDA solution was sprayed directly onto P. patens tissues). P. patens protonemata were grown under continuous white fluorescent light at 25 °C for 24 h. A portion (approximately 2.5 g) of samples (wet P. patens tissue) was ground into powder in liquid nitrogen using a mortar and pestle. The powder was transferred into a solution of 10% trichloroacetic acid and 0.07% 2-mercaptoethanol in acetone and mixed. The suspension was sonicated for 5 min and then incubated for 45 min at −20 °C. After this incubation, the suspension was centrifuged at 9000 g for 20 min at 4 °C. The resulting supernatant was discarded, and the pellet was washed three times with 3 mL of acetone containing 0.07% 2-mercaptoethanol. The final pellet was dried using a vacuum pump. The pellet was resuspended by vortexing for 1 h at 25 °C in 10 mL of lysis buffer consisting of 100 mM Tris-HCl (pH 8.5), 2% SDS, and 50 mM dithiothreitol (DTT). The suspension was then centrifuged at 20,000 g for 20 min at 25 °C. The resulting supernatant was collected as the total protein solution. The concentration of the protein solution was measured using the Lorry method.Citation23)
Digestion of proteins
For the in-solution digestion, 100 μg of protein was subjected to chloroform/methanol extraction.Citation24) The resulting pellet was resuspended with 50 mM NH4HCO3. The solution was reduced with 50 mM DTT and then alkylated with 50 mM iodoacetamide. Proteins were digested using trypsin and lysyl endopeptidase at a 1:100 enzyme/protein ratio at 37 °C for 16 h.
Nanoliquid chromatography-tandem MS analysis
Peptide separation and detection were performed using nanoliquid chromatography tandem mass spectroscopy (nanoLC-MS/MS), which is an Ultimate 3000 nanoLC (Thermo Fisher Scientific, San Jose, CA, USA) and an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific). The system was operated in data-dependent acquisition mode with the installed Xcalibur software (ver. 2.0.7, Thermo Fisher Scientific). The peptides were loaded onto a C18 PepMap trap column (300 μm ID × 5 mm, Thermo Fisher Scientific). The peptides were eluted from the trap column, and separation and spraying were performed using 0.1% formic acid in acetonitrile at a flow rate of 200 μL/min on a C18 Tip column (75 μm ID × 120 mm, Nikkyo Technos, Tokyo, Japan) with a spray voltage of 1.5 kV. Elution was performed with a linear acetonitrile gradient (5–25% in 120 min) in 0.1% formic acid. Full-scan mass spectra were acquired in the Orbitrap over 400–1500 m/z with a resolution of 30,000. A lock mass function was used to obtain high mass accuracy.Citation25) The top ten most intense precursor ions were selected for collision-induced fragmentation in the linear ion trap at a normalized collision energy of 35%. Dynamic exclusion was employed within 90 s to prevent repetitive peptide selection.Citation26)
Protein identification using MASCOT
Acquired MS/MS spectra were subjected to protein identification using MASCOT software (ver. 2.4.1, Matrix Science, London, UK) and the P. patens database (38,480 protein sequences) via Proteome Discoverer (ver. 1.4.0.288, Thermo Fisher Scientific). The P. patens database was obtained from Phytozome database (ver. 9.1, http://www.phytozome.net/). The parameters used in the Mascot searches were as follows: the carbamidomethylation of cysteine was set as a fixed modification; the oxidation of methionine was set as a variable modification; trypsin was specified as the proteolytic enzyme; and one missed cleavage was allowed. The peptide mass tolerance was set at 10 ppm. The fragment mass tolerance was set at 0.8 Da, and the peptide charge was set at +2, +3, and +4. An automatic decoy database search was performed within the search. Mascot results were filtered using the percolator function in Proteome Discoverer to improve the accuracy and sensitivity of peptide identification.Citation27)
Analysis of differential protein abundance using the acquired MS data
For differential analyses, the commercial label-free quantification package SIEVE (ver. 2.1, Thermo Fisher Scientific) was used to compare the relative abundance of peptides and proteins between the control and experimental groups. The chromatographic peaks detected by MS were aligned, and the peptide peaks were detected as frame using the following settings: the frame time width was 5.0 min; the frame m/z width was 10 ppm; and frames were produced on all parent ions subjected to MS/MS scanning. The frames with MS/MS scans were matched to the imported MASCOT results. The MASCOT results were imported with following settings: a minimum peptide ion score of 13 and a maximum percolator q-value of 0.01. The total ion current was used as a normalization factor for the differential analysis.
mRNA expression analysis
Total RNA was extracted from protonemata with the inunuPREP Plant RNA Kit (Analytik Jena Life Science, Jena, Germany). Two microliters of RNA was used for first-strand cDNA synthesis with M-MLV reverse transcriptase (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s instructions. Gene-specific primers were designed according to expressed sequence tag data available for P. patens. The sequences of the primers are listed in Supplemental Table 1. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis was performed following the manufacturer’s protocol (SYBR Premix Ex Taq II, Takara Bio Inc., Shiga, Japan). All RT-PCR reactions were run with three biological replicates on a Thermal Cycler Dice TP800 real-time PCR system (software ver. 3.00D, Takara, Japan). To normalize gene expression, actin (accession no.: AW698983) was used as an internal standard set to 1.0.Citation28)
Results
Protein identification and functional categories of differentially expressed proteins
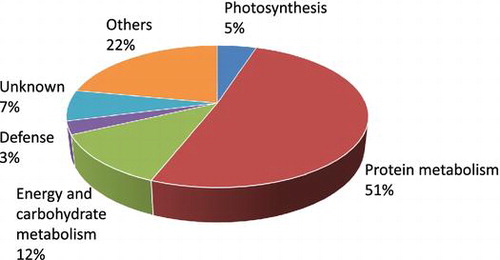
OPDA was shown to suppress the colony growth and rhizoid length of P. patens under a concentration of 10 μM,Citation17) and mechanical stress transiently stimulates OPDA accumulation.Citation19) The morphology of P. patens varies over the course of the life cycle. The protonema and gametophore developmental stages are quite different.Citation29) We examined the effects of OPDA on the growth of P. patens protonemata. P. patens protonemata were incubated in a medium supplemented with OPDA and were then viewed under a microscope (Fig. ). OPDA retarded the growth of protonemata of P. patens in a concentration-dependent manner. When OPDA concentration was more than 10 μM, the protonema growth was clearly inhibited by OPDA. In contrast to OPDA, JA did not show any significant growth-inhibitory effect for P. patens protonemata. Analytical data of endogenous OPDA concentration showed that mechanical wounding transiently elevated OPDA concentration in P. patens protonema in a similar way with higher plants (Fig. ). These results strongly suggest OPDA signaling system functions in protonema and gametophore stages in P. patens.
Fig. 1. Effects of OPDA on protonema prolongation in P. patens.
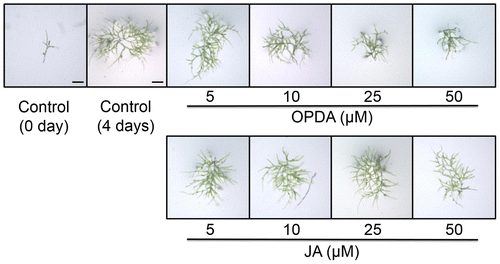
Fig. 2. Accumulation of OPDA in P. patens protonemata after mechanical stress.
A previous report demonstrated that OPDA alters the abundance of proteins involved in light-dependent reactions, the octadecanoid pathway, carbon fixation, glycolysis, and protein synthetic processes in gametophores.Citation19) To examine the effects of OPDA on protein abundance for P. patens protonemata, a proteomic analysis of P. patens protonemata treated with 10 μM OPDA for 24 h was performed. The extracted proteins were digested with trypsin and lysyl endopeptidase, and the resulting peptides were analyzed using nanoLC-MS/MS. The protein levels were compared based on the area under the curve of each matched peptide using SIEVE software; the number of proteins that were matched with more than two peptides was 2662. A subsequent comparative analysis of OPDA-treated protonemata and untreated protonemata revealed that 41 proteins were differentially changed with fold changes of at least 1.5 (p < 0.05) (Table ). The abundance of 40 proteins decreased; only one protein increased in abundance due to OPDA treatment in protonemata.
Table 1. Proteins identified as responsive to OPDA in P. patens protonemata.
Based on their biological properties, these differentially changed proteins were grouped into the following six categories: defense, energy and carbohydrate metabolism, photosynthesis, protein metabolism (proteins synthesis, folding, and degradation), others, and unknown (Fig. ). Twenty-one differentially changed proteins are involved in protein synthesis. One protein is involved in defense, and five proteins are involved in carbohydrate and energy metabolism.
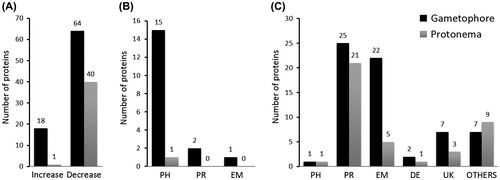
Fig. 3. Comparison of proteins changed in gametophore and protonema.
Proteomic comparison of gametophores and protonemata
We compared the proteome data of gametophoresCitation19) and protonemata to analyze the function of OPDA in P. patens. As shown in Fig. , 82 proteins were differentially expressed in response to OPDA treatment in gametophores (threshold level: 2.5-fold change). The amounts of 41 proteins were differentially altered in protonemata (threshold level: 1.5-fold change). The abundance of most proteins decreased following OPDA treatment in both developmental stages.
The proteins that decreased in abundance in protonemata and gametophores accounted for majority of the proteins that were differently accumulated proteins due to OPDA treatment. Proteins involved in protein metabolism, energy, and carbohydrate metabolism accounted for approximately 65% and 73% of the proteins that decreased in abundance due to OPDA treatment in protonemata and gametophores, respectively. These results suggest that the primary mode of action of OPDA is the repression of protein metabolism and energy consumption processes at both developmental stages. Proteins involved in photosynthesis accounted for 100% and 83% of the proteins that increased abundance due to OPDA treatment in protonemata (one protein) and gametophores, respectively. These results suggest that OPDA would act on photosynthesis in both gametophores and protonemata. The abundance suppression of several histones was notable in P. patens protonemata treated with OPDA.
Proteins with increased abundance due to OPDA treatment of protonemata
The abundance of only one protein, oxygen-evolving enhancer protein 2 (OEE2), was increased by OPDA in protonemata. This protein is encoded by the nuclear genome. OEE2 is required for high levels of O2 evolution. Oxygen-evolving enhancer protein (Pp1s61_321V6.1) belongs to the PsbP family, which is required for increased PSII affinity for the water oxidation site of Cl− and provides the conditions required for high affinity binding to Ca2+.Citation30) Both PsbP and PsbQ are necessary regulators of the biogenesis of optically active PSII. The oxygen-evolving complex (OEC) is responsible for catalyzing the splitting of water to O2 and H+. In flowering plants, such as Arabidopsis, OPDA plays an important role in response to biotic and abiotic stresses.Citation5) OPDA is accumulated in P. patens gametophores due to wounding. OPDA accumulation results in the increased protein abundance in the gametophore stage, including PsbC, psbD, psbE, and allene oxide cyclase (AOC).Citation19) Accordingly, OPDA is likely synthesized to stimulate light-dependent reactions in response to mechanical stress.
Decreased proteins involved in carbohydrate metabolism and energy production
OPDA treatment resulted in the decreased abundance of enzymes involved in carbohydrate metabolism, such as phosphoenolpyruvate carboxykinase, phosphoglucomutase, phosphoglycerate kinase, and pyruvate kinase. Phosphoenolpyruvate carboxykinase (PEPC) is the bottleneck enzyme for gluconeogenesis, which catalyzes the addition of bicarbonate to phosphoenolpyruvate to form oxaloacetate and inorganic phosphate. PEPC plays a crucial role in modulating the balance of carbon metabolism in Arabidopsis.Citation31) Phosphoglucomutase (PGM) catalyzes the interconversion of glucose 1-phosphate and glucose 6-phosphate; the enzyme exists in both plastidial and cytosolic isoforms. The plastidial isoform is essential for transitory starch synthesis in the chloroplasts of leaves, whereas the cytosolic counterpart is essential for glucose phosphate partitioning and the synthesis of sucrose and cell wall components. A lack of PGM (both plastidial and cytosolic isoforms) activities in Arabidopsis resulted in dwarfed growth, premature death, and an inability to develop a functional inflorescence.Citation32) The synthesis of the enzymes phosphoglycerate kinase and pyruvate kinase was suppressed by OPDA; these enzymes are involved in carbon fixation (Calvin cycle) and energy production. Phosphoglycerate kinase is an essential enzyme in the Calvin cycle; it catalyzes the phosphorylation of 3-phosphoglycerate with ATP, which is produced in the light-dependent stage.Citation33) Pyruvate kinase catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to ADP to yield pyruvate and ATP.Citation34)
Proteins with decreased abundance involved in protein metabolism
OPDA suppressed the abundance of ribosomal proteins, transcriptional initiator, and translational inhibitor proteins; these proteins are associated with RNA. Of the 40 proteins with suppressed abundance, over half were involved in protein metabolism (Table , Fig. C). OPDA was shown to down-regulate protein synthesis by suppressing transcription and translation activity. Among the OPDA-repressed proteins involved in protein metabolism, ribosomal proteins were mainly identified in protonemata. The repression of ribosomal protein biosynthesis significantly affects various physiological phenomena in cells. Translational inhibitor proteins (endoribonuclease L-PSP) are active on single-stranded mRNA and inhibit protein synthesis by cleaving mRNA.Citation35,36) When protein synthesis is suppressed in cells, translational inhibitor proteins might be unnecessary. The repression of protein synthesis likely leads to a decreased abundance of translational inhibitor proteins.
Six histones were suppressed by OPDA
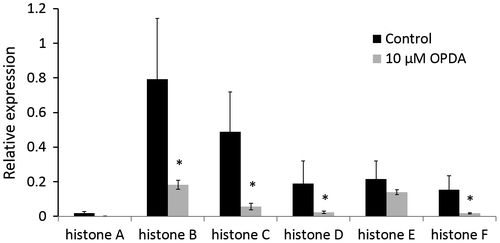
The abundance of six histones was decreased in P. patens protonemata treated with OPDA. To our knowledge, this is the first report that OPDA inhibits histone abundance. These histones all belong to the histone H2A group. Histones are the chief components of chromatin and play an important role in the regulation of gene expression. Histone H2A is important for the packaging of DNA into chromatin; this packaging process is believed to affect gene expression.Citation37) Our results revealed that the abundance of these six histones decreased due to OPDA treatment. Considering the function of histones in gene expression, this finding was worthy of further exploration. We analyzed the expression of these six histone genes at the transcription level by qRT-PCR. As a result, 10 μM OPDA was shown to down-regulate the mRNA expression of these histone genes (Fig. ). These results supported the proteomic data in this study. The qRT-PCR data indicate that OPDA regulates the expression of these histone genes at the transcriptional level. It is likely that OPDA affects cell cycle progression at the protonema stage of P. patens.
Fig. 4. Relative expression of histone genes by qRT-PCR.
Discussion
A comparison of the proteome data from P. patens protonemata treated with OPDA with that from P. patens gametophores treated with OPDA revealed that greater number of proteins were affected in gametophores than in protonemata. Additionally, the magnitude of the changes in protein abundance evoked by OPDA in gametophores was greater that in protonemata. As protonemata differentiate into gametophores, physiological events become more complex; this may explain why OPDA on protein abundance appear to be more significant in gametophores. Whereas OPDA elevates the abundance of a set of proteins in Arabidopsis,Citation12) the abundance of most proteins altered by OPDA decreased in P. patens. Contrary to the changes in protein accumulation due to OPDA observed in Arabidopsis, changes in protein accumulation were restrained in P. patens.
Changes in the abundance of proteins involved in light-dependent reactions were elicited by OPDA in protonemata and gametophores. Accordingly, it was hypothesized that OPDA enhances light-dependent reactions in P. patens. Light-dependent reactions provide oxygen, which is connected to the production of reactive electrophile species (RES). RES and lipid peroxidation appear to be advantageous in plant cells when plants are subjected to stress. RES is presumed to induce the expression of genes related to cell survival.Citation38) As light-dependent reactions are induced by OPDA, redox changes that stimulate signaling cascades to induce the nuclear transcription of mediators may occur.Citation39) OPDA-induced proteins in light-dependent reactions seem to play roles in the stress response at both the protonema and gametophore stages of P. patens. In Arabidopsis, OPDA elevates the accumulation of some proteins involved in photosynthesis, however, the abundance of Rubisco was decreased due to the toxicity of 100 μM OPDA.Citation39) While a high OPDA concentration might give a harmful effect to plants, OPDA treatment resulted in an induction of the synthesis of photosynthesis-related proteins. Taken together, it is likely that the increased abundance of photosynthesis-related proteins due to OPDA treatment is conserved in land plants.
A previous proteomic analysis of P. patens gametophores demonstrated that OPDA treatment induced the abundance of proteins encoded by genes in the chloroplast genome; these proteins were involved in light-dependent reactions.Citation19) In protonemata, no proteins encoded in the chloroplast genome were altered by OPDA treatment. The OPDA-induced accumulation of proteins encoded in the chloroplast genome, which are involved in light-dependent reactions, may be a physiological response of P. patens that is specific to gametophores. The abundance of AOC proteins was induced by OPDA in P. patens gametophores, indicating the presence of positive feedback regulation on OPDA biosynthesis in P. patens gametophores. In contrast, AOC abundance was not increased in P. patens protonemata. Protonema stage is an active growing stage in the life cycle of P. patens, therefore protonemata grow more rapidly than gametophores. Given that OPDA retards growth in P. patens, the positive feedback regulation on OPDA biosynthesis might be suppressed during the protonema stage.
OPDA also reduced the abundance of proteins involved in protein synthesis and carbohydrate metabolism. More than 50% of the affected proteins were involved in protein metabolism (Table , Fig. C). OPDA treatment mainly reduced the abundance of ribosomal proteins in protonemata. Ribosomes are the primary apparatus for biological protein synthesis. The functional repression of ribosomes disrupts the generation of new proteins, thereby arresting the cell growth. As the protonema stage is a period of active growth, more so than the gametophore stage, the growth inhibition observed due to OPDA treatment might be the result of decreased ribosomal protein abundance in P. patens protonemata. Whereas OPDA decreased the abundance of proteins involved in protein metabolism in both protonemata and gametophores, the abundance of additional proteins involved in amino acid synthesis was reduced in gametophores. OPDA likely repressed protein synthesis multilaterally in gametophores.
The abundance of proteins related to carbohydrate metabolism was also suppressed by OPDA treatment in P. patens protonemata; these proteins were not identified in a previous study of gametophores. Given that OPDA synthesis is triggered by wounding, the inhibitory effects of OPDA on the abundance of proteins involved in carbohydrate metabolism have likely been adapted to adverse environmental conditions.
OPDA treatment decreased the accumulation of histones in P. patens protonemata. The decline in histone accumulation might affect DNA replication, as histones are important components of nucleosomes.Citation40). Protonemata grow by apical cell division under the influence of cytokinin; buds are derived from three-faced apical cells that differentiate into gametophores, which contain stem- and leaf-like structures.Citation29) Moreover, cell differentiation is relatively slow in the gametophore stage of P. patens. Protonemata grow more rapidly compared with gametophores. Therefore, suppression of histone expression due to OPDA treatment may retard growth more severely in protonemata than in gametophores.
Genomic analysis revealed that the COI-JAZ system is also present in P. patens. However, JA does not show any significant effect in P. patens. It is likely that OPDA and/or an identified OPDA-related compound binds COI for activation of OPDA signaling in P. patens. Alternatively P. patens might have a unique OPDA signaling system. In either case, considering that OPDA yields physiological effects in land plants, an OPDA signaling system might have been conserved since the emergence of land plants.
In conclusion, OPDA results in the decreased abundance of proteins involved in the metabolism of proteins and carbohydrates at the protonema and gametophore developmental stages. The inhibition of protein synthesis is likely one of the main physiological functions of OPDA in P. patens. This study demonstrated that OPDA suppressed histone expression at both protein level and gene transcription level at the protonema stage. The elucidation of detailed OPDA signaling mechanisms sheds light on P. patens physiology and plant evolution. Our study advances the knowledge of how OPDA regulates physiology in P. patens and increases our understanding of the function of OPDA as a signaling compound in plants.
Author contributions
Study concept and design: Kosaku Takahashi. Acquisition of data: Weifeng Luo, Yohei Nanjo, Setsuko Komatsu, and Kosaku Takahashi. Analysis and interpretation of data: Weifeng Luo, Yohei Nanjo, Setsuko Komatsu, Hideyuki Matsuura, Kosaku Takahashi. Drafting of the manuscript: Weifeng Luo and Kosaku Takahashi. All authors reviewed and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental material for this article can be accessed at http://dx.doi.org/10.1080/09168451.2016.1222268.
TBBB_1222268_Supplementary_Material.docx
Download MS Word (18.7 KB)Acknowledgment
We are grateful to the Chinese Scholarship Council for a scholarship (CSC 201206880001 to W.L.). We thank Dr. Takabayashi at Hokkaido University for his valuable advice.
Notes
Abbreviations: AOC, allene oxide cyclase; AOS, allene oxide synthase; JA, jasmonic acid; LC-MS/MS, liquid chromatography tandem mass spectroscopy; LOX, lipoxygenase; OPDA, 12-oxo-phytodienoic acid; PEPC, phosphoenolpyruvate carboxykinase; PGM, phosphoglucomutase; RES, reactive electrophile species.
References
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007;100:681–697.10.1093/aob/mcm079
- Browse J, Howe GA. New weapons and a rapid response against insect attack. Plant Physiol. 2008;146:832–838.10.1104/pp.107.115683
- Howe GA, Jander G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008;59:41–66.10.1146/annurev.arplant.59.032607.092825
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009;60:183–205.10.1146/annurev.arplant.043008.092007
- Böttcher C, Pollmann S. Plant oxylipins: plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009;276:4693–4704.10.1111/j.1742-4658.2009.07195.x
- Schaller A, Stintzi A. Enzymes in jasmonate biosynthesis – structure, function, regulation. Phytochemistry. 2009;70:1532–1538.10.1016/j.phytochem.2009.07.032
- Anterola A, Göbel C, Hornung E, et al. Physcomitrella patens has lipoxygenases for both eicosanoid and octadecanoid pathways. Phytochemistry. 2009;70:40–52.10.1016/j.phytochem.2008.11.012
- Bandara PKGSS, Takahashi K, Sato M, et al. Cloning and functional analysis of an allene oxide synthase in Physcomitrella patens. Biosci. Biotechnol. Biochem. 2009; 73: 2356–2359.
- Hashimoto T, Takahashi K, Sato M, et al. Cloning and characterization of an allene oxide cyclase, PpAOC3, in Physcomitrella patens. Plant Growth Regul. 2011;65:239–245.10.1007/s10725-011-9592-z
- Stumpe M, Göbel C, Faltin B, et al. The moss Physcomitrella patens contains cyclopentenones but no jasmonates: mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 2010;188:740–749.10.1111/j.1469-8137.2010.03406.x
- Balbi V, Devoto A. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 2008;177:301–318.
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005;139:1268–1283.10.1104/pp.105.067058
- Goetz S, Hellwege A, Stenzel I, et al. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiol. 2012;158:1715–1727.10.1104/pp.111.192658
- Dave A, Hernández ML, He Z, et al. 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell. 2011;23:583–599.10.1105/tpc.110.081489
- Rensing SA, Lang D, Zimmer AD, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69.10.1126/science.1150646
- Hiss M, Laule O, Meskauskiene RM, et al. Large-scale gene expression profiling data for the model moss Physcomitrella patens aid understanding of developmental progression, culture and stress conditions. Plant J. 2014;79:530–539.10.1111/tpj.12572
- Ponce De León I, Schmelz EA, Gaggero C, et al. Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 2012;13:960–974.10.1111/mpp.2012.13.issue-8
- Scholz J, Brodhun F, Hornung E, et al. Biosynthesis of allene oxides in Physcomitrella patens. BMC Plant Biol. 2012;12:228. doi:10.1186/s12870-016-0865-610.1186/1471-2229-12-228
- Toshima E, Nanjo Y, Komatsu S, et al. Proteomic analysis of Physcomitrella patens treated with 12-oxo-phytodienoic acid, an important oxylipin in plants. Biosci. Biotechnol. Biochem. 2014;78:946–953.10.1080/09168451.2014.912112
- Ashton NW, Cove DJ. Isolation and preliminary characterization of auxotrophic and analog resistant mutants of moss, Physcomitrella patens. Mol. Gen. Gen. 1977;154:87–95.10.1007/BF00265581
- Kajiwara A, Abe T, Hashimoto T, et al. Efficient synthesis of (+)- cis-12-oxo-phytodienoic acid by an in vitro enzymatic reaction. Biosci. Biotechnol. Biochem. 2012;76:2325–2328.10.1271/bbb.120506
- Yamamoto Y, Ohshika J, Takahashi T, et al. Functional analysis of allene oxide cyclase, MpAOC, in the liverwort Marchantia polymorpha. Phytochemistry. 2015;116:48–56.10.1016/j.phytochem.2015.03.008
- Komatsu S, Han C, Nanjo Y, et al. Label-free quantitative proteomic analysis of abscisic acid effect in early-stage soybean under flooding. J. Proteome Res. 2013;12:4769–4784.10.1021/pr4001898
- Nanjo Y, Skultety L, Uváčková L, et al. Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J. Proteome Res. 2012;11:372–385.10.1021/pr200701y
- Olsen JV, de Godoy LMF, Li GQ, et al. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 2005;4:2010–2021.10.1074/mcp.T500030-MCP200
- Zhang Y, Wen Z, Washburn MP, et al. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal. Chem. 2009;81:6317–6326.10.1021/ac9004887
- Brosch M, Yu L, Hubbard T, et al. Accurate and sensitive peptide identification with Mascot Percolator. J. Proteome Res. 2009;8:3176–3181.10.1021/pr800982s
- Le Bail A, Scholz S, Kost B. Evaluation of reference genes for RT qPCR analyses of structure-specific and hormone regulated gene expression in Physcomitrella patens gametophytes. PLoS ONE. 2013;8:e70998.10.1371/journal.pone.0070998
- Roberts AW, Roberts EM, Haigler CH. Moss cell walls: structure and biosynthesis. Front Plant Sci. 2012;3:1–7.
- Kochhar A, Khurana JP, Tyagi AK. Nucleotide sequence of the psbP gene encoding precursor of 23-kDa polypeptide of oxygen-evolving complex in Arabidopsis thaliana and its expression in the wild-type and a constitutively photomorphogenic mutant. DNA Res. 1996;3:277–285.10.1093/dnares/3.5.277
- Shi J, Yi K, Liu Y, et al. Phosphoenolpyruvate carboxylase in Arabidopsis leaves plays a crucial role in carbon and nitrogen metabolism. Plant Physiol. 2015;167:671–681.10.1104/pp.114.254474
- Malinova I, Kunz HH, Alseekh S, et al. Reduction of the cytosolic phosphoglucomutase in Arabidopsis reveals impact on plant growth, seed and root development, and carbohydrate partitioning. PLoS ONE. 2014;9:e112468.10.1371/journal.pone.0112468
- Joshi R, Karan R, Singla-Pareek SL, et al. Ectopic expression of Pokkali phosphoglycerate kinase-2 (OsPGK2-P) improves yield in tobacco plants under salinity stress. Plant Cell Rep. 2016;35:27–41.10.1007/s00299-015-1864-z
- Romano AH, Conway T. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 1996;147:448–455.10.1016/0923-2508(96)83998-2
- Morishita R, Kawagoshi A, Sawasaki T, et al. Ribonuclease activity of rat liver perchloric acid-soluble Protein, a potent inhibitor of protein synthesis. J. Biol. Chem. 1999;274:20688–20692.10.1074/jbc.274.29.20688
- Hedegaard J, Hauge M, Fage-Larsen J, et al. Investigation of the translation-initiation factor IF2 gene, infB, as a tool to study the population structure of Streptococcus agalactiae. Microbiology-UK. 2000;146:1661–1670.10.1099/00221287-146-7-1661
- Marino-Ramirez L, Jordan IK, Landsman D. Multiple independent evolutionary solutions to core histone gene regulation. Genome Biol. 2006;7:1–17.
- Farmer EE, Mueller MJ. ROS-mediated lipid peroxidation and RES-activated signaling. Annu. Rev. Plant Biol. 2013;64:429–450.10.1146/annurev-arplant-050312-120132
- Dueckershoff K, Mueller S, Mueller MJ, et al. Impact of cyclopentenone-oxylipins on the proteome of Arabidopsis thaliana. Biochim. Biophys. Acta. 2008;1784:1975–1985.10.1016/j.bbapap.2008.09.003
- Kornberg RD. Structure of chromatin. Annu. Rev. Biochem. 1977;46:931–954.10.1146/annurev.bi.46.070177.004435
