Abstract
To date, the majority of molecular genetic studies in algae have utilized a fairly limited range of markers such as the plastid rbcL gene and spacer, the mitochondrial cox2-3 spacer or the nuclear ribosomal DNA and spacers. The lack of available markers has been particularly problematic in studies of within-species variation. Whilst microsatellites are now being developed in many algal species, there remains a need for universal markers that can be applied to a wide range of species. The increasing availability of complete plastid genome sequences for several algae has allowed us to develop two sets of universal primers, similar to those available in higher plants, for the amplification of coding and non-coding regions of the plastid genome in red and green algae. These markers are expected to be useful in a broad range of algal population genetic and phylogenetic studies.
Introduction
The development of universal primers for the amplification of various regions of the chloroplast genome, particularly of non-coding DNA, has provided invaluable tools for evolutionary genetic studies in plants (Taberlet et al., Citation1991; Demesure et al., Citation1995; Dumolin-Lapegue et al., Citation1997). The original paper, describing three pairs of consensus primers for the amplification of the trnT – trnL intergenic region, the trnL intron and the trnL-trnF intergenic region in plant taxa ranging from bryophytes to angiosperms (Taberlet et al., Citation1991), has now been cited over 500 times. These universal plastid primers are routinely used in plant phylogenetic studies and particularly for population genetics since many of these regions, particularly the trnT – L – F intergenic regions and intron, display higher levels of sequence variation than more traditional markers such as the RuBisCO large subunit gene (rbcL; e.g. McDade & Moody, Citation1999).
In algae, phylogenetic studies have generally utilized either the rbcL gene of the plastid genome (e.g. Freshwater et al., Citation1994; McIvor et al., Citation2002) or the nuclear ribosomal DNA small subunit (SSU), and recently also some more variable domains of the large subunit (LSU; e.g. Harper & Saunders, Citation2001, Citation2002). For evolutionary studies at a finer scale, the lack of tools like the plant universal primers has been a handicap. The majority of phylogeographic studies have employed the internal transcribed spacers (ITS) in the ribosomal cistron of the nuclear genome (for review see Wattier & Maggs, Citation2001). The popularity of the ITS region can be attributed to the relatively high rate of nucleotide substitution, permitting comparison of relatively recently diverged taxa. In addition the ITS region can be readily PCR-amplified and sequenced with conserved primers positioned in the cistronic regions. However, many of the earlier studies used the suite of primers published by White et al. (Citation1990) for fungi, and their lack of specificity often resulted in the amplification of spurious algal sequences (such as those investigated by Bown et al., Citation2003). Furthermore, the occurrence of intraindividual variation in ITS sequences means that cloning is often required prior to sequencing (Famà et al., Citation2000; Lange et al., Citation2002). Single-locus nuclear microsatellites have now been employed in population studies in red, green and brown algae, providing information on kinship, paternity and gene flow (e.g. van der Strate et al., Citation2002; Engel et al., Citation2002; Coyer et al., Citation2003). Their development has been very laborious, and in most cases they can be used only in the species for which they were defined, or for very closely related algae.
Two non-coding organellar DNA regions are widely used in algae. In many red and brown algae, the spacer between the genes for the large and small subunits of RuBisCo (rbcL – rbcS spacer) can be amplified using primers originally designed for members of the Gracilariaceae and Gigartinales (Destombe & Douglas, Citation1991; Maggs et al., Citation1992). This marker has been developed as a target for PCR – SSCP (single-stranded conformational polymorphism) (Zuccarello et al., Citation1999). However, the RuBisCo spacer is short (typically 75 – 120 bp), limiting its value. For red algae, the mitochondrial cox2-3 spacer amplified by the primers of Zuccarello et al. (Citation1999) is a potentially universal marker for non-coding DNA. The spacer, approximately 300 – 350 bp long, has proved informative in a range of systematic and phylogeographic studies in red algae (Gabrielsen et al., Citation2002; Marston and Villalard-Bohnsack, Citation2002; Zuccarello & West, Citation2002; Zuccarello et al., Citation2002).
Although the various markers mentioned above have proved to be informative in particular cases, often they are not appropriate e.g. they do not provide enough resolution (at the population level) or they are too variable and approaching saturation (at higher taxonomic levels). Increasingly, therefore, phylogenetic and phylogeographic studies require two or more sets of data, aimed at resolving different depths of evolutionary relationships (e.g. Bellorin et al., Citation2002; Draisma et al., Citation2002; Hayden et al., Citation2003).
There are several reasons why universal primers similar to the ones described for plants have not been developed for algae. Firstly, comparison of the available algal plastid genome sequences (see below) shows that they display high levels of structural and sequence diversity compared with plants, even within the divisions/phyla Chlorophyta (green algae) and Rhodophyta (red algae). Furthermore, such comparisons also show that there tends to be a lower percentage of intergenic/intronic DNA in algal genomes, particularly in the highly compact plastid genomes of rhodophytes (with certain exceptions e.g. the rbcL – rbcS spacer). Finally, the availability of algal plastid sequence data is limited: until fairly recently, the only complete plastid genome sequences available were those of the rhodophyte Porphyra purpurea (Reith & Munholland, Citation1995), the glaucocystophyte Cyanophora paradoxa (Stirewalt et al., Citation1995), the heterokont Odontella sinensis (Kowallik et al., Citation1995) and the chlorophyte Chlorella vulgaris (Wakasugi et al., Citation1997).
Attempts to use the plant universal chloroplast primers in algae have met with extremely limited success – a few work in green algae (Taberlet et al., Citation1991) but none have been successfully amplified in red algae due to the degree of divergence between streptophytes and rhodophytes and high levels of plastid genome rearrangement (Ohta et al., Citation1997; Wattier et al., Citation2001). Advances in DNA sequencing methodologies and sequence analysis have led to a vast increase in information available in DNA sequence databanks. The establishment of genome sequencing projects in a variety of organisms has resulted in a proliferation of complete chloroplast genome sequences from a range of plants and algae including the rhodophytes Cyanidium caldarium (Glockner et al., Citation2000) and Cyanidioschyzon merolae (Ohta et al., Citation2003), the chlorophytes Nephroselmis olivacea (Turmel et al., Citation1999) and Mesostigma viride (Lemieux et al., Citation2000), the charophyte Chaetosphaeridium globosum (Turmel et al., Citation2002) and the cryptophyte Guillardia theta (Douglas & Penny, Citation1999). Consequently, it is now possible to use comparative genomics to develop novel tools for molecular analyses in algae. We have utilized such an approach to develop two sets of universal primers for the amplification of coding and non-coding sections of the plastid genome in red and green algae.
Materials and methods
Primer design
Since the plastid genomes of red and green algae are so divergent (Ohta et al., Citation1997), it is unlikely that truly universal primers could be developed that would amplify in both Rhodophyta and Chlorophyta. We therefore decided to design separate sets of primers for each phylum. Universal primers for rhodophytes were designed by a comparative analysis of the complete plastid genome sequences of Porphyra purpurea (Roth) C. Agardh (GenBank accession number U38804; Reith & Munholland, Citation1995), Cyanidium caldarium Geitler (AF022106; Glockner et al., Citation2000) and Cyanidioschyzon merolae De Luca, Taddei et Varano (AB005283; Ohta et al., Citation2003). For chlorophytes, the complete plastid sequences of Chlorella vulgaris (AB001684; Wakasugi et al., Citation1997), Nephroselmis olivacea Stein (AF137379; Turmel et al., Citation1999), Mesostigma viride Lauterborn (AF166114; Lemieux et al., Citation2000) and Chaetosphaeridium globosum (Nordstedt) Klebahn (AF494278; Turmel et al., Citation2002) were used. A search for syntenic regions of the genomes revealed that the cluster of genes containing mainly ribosomal proteins between rps10 and dnaK was conserved across all three rhodophytes () and that the gene cluster between rps11 and rpl2 was conserved in chlorophytes (). Sequences of individual genes were edited using the ASSEMBLE program (Genetics Computer Group, Wisconsin, USA) and aligned using the BioEdit software package (V5.0.9; www.mbio.ncsu.edu/BioEdit/bioedit.html). For rhodophytes, nine overlapping pairs of primers designed to amplify the complete cluster of 30 genes (13 – 17 kb) are shown in . For chlorophytes, eight pairs of primers () were designed to amplify the cluster of 10 – 12 genes (∼5 kb).
Fig. 1. Diagram showing the universal rhodophyte primer (URP) pairs designed to amplify the 13 – 15 kb rps10 – dnaK ribosomal protein gene cluster conserved across Porphyra purpurea, Cyanidium caldarium and Cyanidioschyzon merolae (not shown to scale).
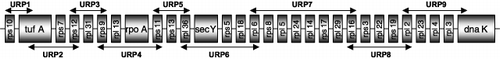
Fig. 2. Diagram showing the universal chlorophyte primer (UCP) pairs designed to amplify the ∼5 kb rps11 – rpl2 gene cluster conserved across Nephroselmis olivacea, Chlorella vulgaris, Mesostigma viride and Chaetosphaeridium globosum (not shown to scale). (a) Primer pair UPC3 also amplifies ORF54 which lies between rps8 and rpl5 in Chlorella vulgaris. (b) Primer pair UPC7 also amplifies the rps22 gene which lies between rps3 and rps19 in Mesostigma viride and Chaetosphaeridium globosum. (c) Primer pair UPC8 also amplifies ORF45 which lies between rps19 and rpl2 in Chlorella vulgaris.
Table 1. Universal rhodophyte primers (see for details of loci)
Table 2. Universal chlorophyte primers (see for details of loci)
Primer utility
To assess the cross-species utility of the rhodophyte primers, they were tested on the following species: Porphyra purpurea (Bangiales), Rhodochorton purpureum (Lightfoot) Rosenvinge (Acrochaetiales), Phycodrys rubens (L.) Batters (Ceramiales), Corallina officinalis L. (Corallinales), Gelidium pulchellum (Turner) Kützing (Gelidiales), Mastocarpus stellatus (Stackhouse) Guiry (Gigartinales), Palmaria palmata (L.) O. Kuntze (Palmariales) and Plocamium cartilagineum (L.) P. Dixon (Plocamiales). The chlorophyte primers were tested on Dunaliella salina Teodoresco (Chlorophyceae), Ulva lactuca L. (Ulvophyceae), Codium fragile (Suringar) Hariot (Ulvophyceae) and Coleochaete orbicularis Pringsheim (Charophyceae). Total DNA was extracted either by the phenol-chloroform protocol of Wattier et al. (Citation2000) or using DNeasy Mini-Preps (Quiagen, Hilden). PCR was carried out on a MWG Primus thermal cycler using the following parameters: initial denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 48 – 56°C (see and for optimum annealing temperatures) for 1 min, extension at 72°C for 1 – 5 min (1 min per kb of expected sized product) and a final extension at 72°C for 5 min. PCR was carried out in a total volume of 10 μl containing 100 ng DNA, 5 pmol of forward and reverse primers, 1 × PCR reaction buffer (5 mM Tris-HCl [pH9.1], 1.6 mM [NH4]2SO4, 15 μg μl−1 BSA), 2.5 mM MgCl2 and 1 U Taq polymerase (Genetix). PCR products were resolved on 2% agarose gels and visualised by ethidium bromide staining.
Results and discussion
As recently as July 2001, Wattier et al. (Citation2001) stated that ‘… the evolutionary diversity of the Rhodophyta is so great that the possibility of designing universal primers for them is very limited.’ Thanks to recent advances in whole-genome sequencing, comparative approaches now offer real scope for developing truly universal markers in both red and green algae. We have used such an approach to develop a suite of primers to amplify the 13 – 17 kb rps10-dnaK gene cluster in all rhodophytes and another to amplify the 5 kb rps11-rpl2 cluster in chlorophytes. The new primers reported here represent much-needed additional tools for population and evolutionary studies in both red and green algae.
The amplification success of primer pairs varied between 50% and 100% in the species tested in this study and generated PCR products of the expected size, based on the previously published complete plastid genome sequences ( and ). As with all universal primers, some degree of largely empirical work is necessary to optimize reaction conditions in different taxa, particularly with those primers that amplify larger PCR products. Under the correct conditions, however, most of the primer pairs amplified a single band in each species studied (see and ). Whilst it may take some effort to optimise PCR conditions for all primers, it is possible to use them to amplify a product in the species under study, sequence the product and then develop potentially more robust species-specific or clade-specific primers for subsequent analyses (e.g. to minimise the problem of amplifying products from epiphytic algae or, conversely, the hosts of parasites or endophytes). Furthermore, since the universal chlorophyte primers tend to amplify shorter products between contiguous genes, it is possible to ‘skip’ a gene and use a combination of primers (e.g. use the forward primer for UCP1 and the reverse primer for UCP2) to amplify larger regions, particularly if it proves difficult to optimize a specific pair of primers for the alga(e) in question.
Table 3. Cross species amplification of rhodophyte universal primers. Ppu – Porphyra purpure a; Rpu – Rhodochorton purpureu m; Pru – Phycodrys ruben s; Cof – Corallina officinali s; Gpu – Gelidium pulchellu m; Mst – Mastocarpus stellatu s; Ppa – Palmaria palmat a; Pca – Plocamium cartilagineu m
Table 4. Cross species amplification of chlorophyte universal primers. Dsa – Dunaliella salin a; Ula – Ulva lactuc a; Cfr – Codium fragil e; Cor – Coleochaete orbiculari s
The various regions amplified by these primers display a range of levels of sequence divergence between the species from which they were designed. Pairwise sequence identities ranged from 40% (UCP7, Nephroselmis olivacea vs Chaetosphaeridium globosum and Chlorella vulgaris vs Chaetosphaeridium globosum) to 69% (UCP2, Mesostigma viride vs Chaetosphaeridium globosum; and ), suggesting that different primer pairs can be used to provide the desired degree of resolution appropriate for the taxonomic level under study. For example, less conserved loci may be useful for population level studies whereas the primers that amplify the more conserved regions would be better suited to analysis at the specific or generic level. Since the amount of non-coding DNA amplified by each primer pair varies greatly across taxa, however, it is not feasible to calculate levels of divergence separately for coding and non-coding regions.
Table 5. Pairwise sequence similarity by locus for rhodophyte universal primers
Table 6. Pairwise similarity by locus for chlorophyte universal plastid primers (N. o. – Nephroselmis olivacea; C. v. – Chlorella vulgaris; M. v. – Mesostigma viride; C. g. – Chaetosphaeridium globosum.)
Although a gene cluster identical to that found in rhodophytes is also present in the secondary plastid genomes of Odontella sinensis and Guillardia theta, higher levels of sequence divergence between rhodophytes and those algae with secondary red plastids (including heterokonts, haptophytes and cryptophytes; Stoebe & Kowallik, Citation1999) mean that it is unlikely that these primers will amplify as successfully in these taxa as as in rhodophytes. Because of this, it may be feasible to use a similar approach based on the plastid genome sequences of Porphyra purpurea, Cyanidium caldarium, Odontella sinensis and Guillardia theta to design a comparable set of primers for use in lineages that acquired red-type plastids via secondary endosymbiotic events. Interestingly, examination of the plastid genome of the glaucocystophyte Cyanophora paradoxa reveals that the same cluster appears to have been ‘split’ into four and redistributed around the genome but, again, it is unlikely that our primers will amplify in glaucocystophytes.
In summary, we have developed a novel suite of primers that provide potentially valuable tools for both population and phylogenetic studies in red and green algae. As the existing repertoire of markers is extremely limited, the effort of optimising universal primers in individual studies is greatly outweighed by the benefits of having new markers, particularly where existing ones were unsuitable.
Acknowledgements
The authors would like to thank Joe Zuccarello and Wytze Stam for helpful comments on an earlier version of the manuscript. This work was funded under the EU Framework V contract ALIENS.
References
References
- Bellorin , AM , Oliveira , MC and Oliveira , EC . 2002 . Phylogeny and systematics of the marine algal family Gracilariaceae (Gracilariales, Rhodophyta) based on small subunit rDNA and ITS sequences of Atlantic and Pacific species . J. Phycol , 38 : 551 – 563 .
- Bown , P , Plumb , J , Sanchez-Baracaldo , P , Hayes , PK and Brodie , J . 2003 . Sequence heterogeneity of green (Chlorophyta) endophytic algae associated with a population of Chondrus crispus (Gigartinaceae, Rhodophyta) . Eur. J. Phycol , 38 : 153 – 163 .
- Coyer , JA , Peters , AF , Stam , WT and Olsen , JL . 2003 . Post-ice age recolonization and differentiation of Fucus serratus L. (Phaeophyceae; Fucaceae) populations in Northern Europe . Mol. Ecol , 12 : 1817 – 1829 .
- Demesure , B , Sodzi , N and Petit , RJ . 1995 . A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants . Mol. Ecol , 4 : 129 – 131 .
- Destombe , C and Douglas , SE . 1991 . Rubisco spacer sequence divergence in the rhodophyte alga Gracilaria verrucosa and closely related species . Curr. Genet , 19 : 395 – 398 .
- Douglas , SE and Penny , SL . 1999 . The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae . J. Mol. Evol , 48 : 236 – 244 .
- Draisma , SGA , Olsen , JL , Stam , WT and Prud'Homme van Reine , WF . 2002 . Phylogenetic relationships within the Sphacelariales (Phaeophyceae): rbcL, RUBISCO spacer and morphology . Eur. J. Phycol , 37 : 385 – 402 .
- Dumolin-Lapegue , S , Pemonge , MH and Petit , RJ . 1997 . An enlarged set of consensus primers for the study of organelle DNA in plants . Mol. Ecol , 6 : 393 – 397 .
- Engel , CR , Valero , M , Lagadeuc , Y and Destombe , C . 2002 . Non-random mating in controlled multiple-donor crosses in Gracilaria gracilis (Gracilariaceae, Rhodophyta) . Eur. J. Phycol , 37 : 179 – 190 .
- Famà , P , Olsen , JL , Stam , WT and Procaccini , G . 2000 . High levels of intra- and inter-individual polymorphism in the rDNA ITS1 of Caulerpa racemosa (Chlorophyta) . Eur. J. Phycol , 35 : 349 – 356 .
- Freshwater , DW , Fredericq , S , Butler , BS , Hommersand , MH and Chase , MW . 1994 . A gene phylogeny of the red algae (Rhodophyta) based on plastid rbcL . Proc. Natl. Acad. Sci. USA , 91 : 7281 – 7285 .
- Gabrielsen , TM , Brochmann , C and Rueness , J . 2002 . The Baltic Sea as a model system for studying postglacial colonization and ecological differentiation, exemplified by the red alga Ceramium . Mol. Ecol , 11 : 2083 – 2095 .
- Glockner , G , Rosenthal , A and Valentin , K . 2000 . The structure and gene repertoire of an ancient red algal plastid genome . J. Mol. Evol , 51 : 382 – 390 .
- Harper , JT and Saunders , GW . 2001 . Molecular systematics of the Florideophyceae (Rhodophyta) using nuclear large and small subunit rDNA sequence data . J. Phycol , 37 : 1073 – 1082 .
- Harper , JT and Saunders , GW . 2002 . A re-classification of the Acrochaetiales based on molecular and morphological data, and establishment of the Colaonematales ord. nov. (Florideophyceae, Rhodophyta) . Eur. J. Phycol , 37 : 463 – 476 .
- Hayden , HS , Blomster , J , Maggs , CA , Silva , PC , Stanhope , MJ and Waaland , JR . 2003 . Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera . Eur. J. Phycol , 38 : 277 – 294 .
- Kowallik , KV , Stoebe , B , Schaffran , I , Kroth-Pancic , P and Freier , U . 1995 . The chloroplast genome of a chlorophyll a + c- containing alga, Odontella sinensis . Plant. Mol. Biol. Rep , 13 : 336 – 342 .
- Lange , M , Chen , YQ and Medlin , LK . 2002 . Molecular genetic delineation of Phaeocystis species (Prymnesiophyceae) using coding and non-coding regions of nuclear and plastid genomes . Eur. J. Phycol , 37 : 77 – 92 .
- Lemieux , C , Otis , C and Turmel , M . 2000 . Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution . Nature , 403 : 649 – 652 .
- Maggs , CA , Douglas , SE , Fenety , J and Bird , CJ . 1992 . A molecular and morphological analysis of the Gymnogongrus devoniensis complex in the North Atlantic . J. Phycol , 28 : 214 – 232 .
- Marston , M and Villalard-Bohnsack , M . 2002 . Genetic variability and potential sources of Grateloupia doryphora (Halymeniaceae, Rhodophyta), an invasive species in Rhode Island waters (USA) . J. Phycol , 38 : 649 – 658 .
- McDade , LA and Moody , ML . 1999 . Phylogenetic relationships among Acanthaceae: Evidence from noncoding trnL-trnF chloroplast DNA sequences . Am. J. Bot , 86 : 70 – 80 .
- McIvor , L , Maggs , CA and Stanhope , MJ . 2002 . rbcL sequences indicate a single evolutionary origin of multinucleate cells in the red algal tribe Callithamnieae . Mol. Phylog. Evol , 23 : 433 – 446 .
- Ohta , N , Sato , N , Nozaki , H and Kuroiwa , T . 1997 . Analysis of the cluster of ribosomal protein genes in the plastid genome of a unicellular red alga Cyanidioschyzon merolae: Translocation of the str cluster as an early event in the rhodophyte-chromophyte lineage of plastid evolution . J. Mol. Evol , 45 : 688 – 695 .
- Ohta , N , Matsuzaki , M , Misumi , O , Miyagishima , S , Nozaki , H , Tanaka , K , Shin-i , T , Kohara , Y and Kuroiwa , T . 2003 . Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae . DNA Res , 10 : 67 – 77 .
- Reith , ME and Munholland , J . 1995 . Complete nucleotide sequence of the Porphyra purpurea chloroplast genome . Plant Mol. Biol. Rep , 13 : 333 – 335 .
- Stirewalt , VL , Michalowski , CB , Löffelhardt , W , Bohnert , HJ and Bryant , DA . 1995 . Nucleotide sequence of the cyanelle genome from Cyanophora paradoxa . Plant Mol. Biol. Rep , 13 : 327 – 332 .
- Stoebe , B and Kowallik , KV . 1999 . Gene-cluster analysis in chloroplast genomics . Trends in Genetics , 15 : 344 – 347 .
- Taberlet , P , Gielly , L , Patou , G and Bouvet , J . 1991 . Universal primers for amplification of three non-coding regions of chloroplast DNA . Plant Mol. Biol , 17 : 1105 – 1109 .
- Turmel , M , Otis , C and Lemieux , C . 1999 . The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes . Proc. Natl. Acad. Sci. USA , 96 : 10248 – 10253 .
- Turmel , M , Otis , C and Lemieux , C . 2002 . The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants . Proc. Natl. Acad. Sci. USA , 99 : 11275 – 11280 .
- van der Strate , HJ , Boele-Bos , SA , Olsen , JL , van de Zande , L and Stam , WT . 2002 . Phylogeographic studies in the tropical seaweed Cladophoropsis membranacea (Chlorophyta, Ulvophyceae) reveal a cryptic species complex . J. Phycol , 38 : 572 – 582 .
- Wakasugi , T , Nagai , T , Kapoor , M , Sugita , M , Ito , M , Ito , S , Tsudzuki , J , Nakashima , K , Tsudzuki , T , Suzuki , Y , Hamada , A , Ohta , T , Inamura , A , Yoshinaga , K and Sugiura , M . 1997 . Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: the existence of genes possibly involved in chloroplast division . Proc. Natl. Acad. Sci. USA , 94 : 5967 – 5972 .
- Wattier , R , Prodohl , PA and Maggs , CA . 2000 . DNA extaction protocol for red seaweeds . Plant Mol. Biol. Rep , 18 : 275 – 281 .
- Wattier , RA and Maggs , CA . 2001 . Intraspecific variation in seaweeds: the application of new tools and approaches . Adv. Bot. Res , 35 : 171 – 212 .
- Wattier , RA , Davidson , AL , Ward , BA and Maggs , CA . 2001 . cpDNA-RFLP in Ceramium (Rhodophyta): Intraspecific polymorphism and species-level phylogeny . Am. J. Bot , 88 : 1209 – 1213 .
- White TJ Bruns T Lee S Taylor J 1990 Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In PCR Protocols: a Guide to Methods and Applications (Innis, M., Gelfand, J., Sninsky, J. & White, T.J., editors) 315 322 Academic Press, Orlando, Florida
- Zuccarello , GC , Burger , G , West , JA and King , RJ . 1999 . A mitochondrial marker for red algal intraspecific relationships . Mol. Ecol , 8 : 1443 – 1447 .
- Zuccarello , GC and West , JA . 2002 . Phylogeography of the Bostrychia calliptera-B. pinnata complex (Rhodomelaceae, Rhodophyta) and divergence rates based on nuclear, mitochondrial and plastid DNA markers . Phycologia , 41 : 49 – 60 .
- Zuccarello , GC , Sandercock , B and West , JA . 2002 . Diversity within red algal species: variation in world-wide samples of Spyridia filamentosa (Ceramiaceae) and Murrayella periclados (Rhodomelaceae) using DNA markers and breeding studies . Eur. J. Phycol , 37 : 403 – 417 .