ABSTRACT
In this study we investigated Ralfsia-like crusts (i.e. excluding Ralfsia sensu stricto, Stragularia spp. and/or Scytosiphonaceae crustose phases) with an emphasis on the North Atlantic Ocean using molecular data (COI-5P, ITS and rbcL-3P) combined with morpho-anatomical comparisons of type material and contemporary specimens. Of the four species of Ralfsia previously reported in Europe, only R. fungiformis is presently recognized as belonging to Ralfsia sensu stricto, Ralfsiaceae, with the remaining species, R. lucida, R. ovata and R. verrucosa, of uncertain taxonomic status. Our study revealed 11 independent genetic lineages of Ralfsia-like taxa, which were not assignable to any of the recognized families of the Ralfsiales. To accommodate this diversity, we propose Pseudoralfsiaceae Parente, Fletcher & G.W.Saunders fam. nov., including two new genera Pseudoralfsia Parente, Fletcher & G.W.Saunders gen. nov. and Nuchella Parente, Fletcher & G.W.Saunders gen. nov. The first genus includes Pseudoralfsia verrucosa (Areschoug) Parente, Fletcher & G.W.Saunders comb. nov. as the generitype (the only species of the three remaining European species of uncertain taxonomic status assigned to Ralfsia that was reassessed here), P. azorica Parente, Fletcher & G.W.Saunders sp. nov. and seven undescribed genetic groups, which require additional vouchers for description. The second genus has as generitype Nuchella vesicularis Parente, Fletcher & G.W.Saunders sp. nov., and also includes N. sp._1MP, which was represented by a single sterile specimen from Swanage, UK and consequently not characterized. Three characteristics differentiate Nuchella from Pseudoralfsia – the former typically with numerous vesicles, non-synchronous development of the plurangia and hair pits arising from both middle and lower cells of the erect filaments. Species of Pseudoralfsiaceae can be distinguished from Ralfsia sensu stricto (Ralfsiaceae) mainly by DNA sequences and by consistently having frequent hair pits, and typically unsymmetrical thalli.
Introduction
Systematics of the brown algal order Ralfsiales (Phaeophyceae), proposed by Nakamura (Citation1972), are unstable. Long considered nomenclaturally invalid and representing an artificial grouping, Ralfsiales was later validated and amended by Lim et al. (Citation2007) based on Rubisco large subunit gene (rbcL) sequence data. Within the order, these authors recognized the family Ralfsiaceae as comprising the genera Analipus, Endoplura, Heteroralfsia and Ralfsia, and proposed a new family Neoralfsiaceae to accommodate the new genus Neoralfsia, which was based on Ralfsia expansa (J.Agardh) J.Agardh. They also recognized the family Mesosporaceae, but with some hesitation probably owing to the lack of the generitype in their analyses. Finally, they also revealed the presence of a separate clade within the order, which they named ‘?Ralfsia?’, that was genetically distant from members of all the other families and based on two unidentified specimens informally referred to Ralfsia. Subsequent molecular studies justified recognition of Mesosporaceae by including the generitype Mesospora schmidtii Weber-van Bosse, while at the same time providing a better understanding of several other known and novel species of Mesospora (Poong et al., Citation2013, Citation2014, Citation2017). Most recently Léon-Alvaréz et al. (Citation2017) proposed the new family Hapalospongidiaceae based on the generitype Hapalospongidion gelatinosum De A.Saunders while Parente & Saunders (Citation2019) published a morphological and molecular evaluation of Ralfsia sensu stricto, Ralfsiaceae in Canada. Of the species previously assigned to this genus, only the type species R. fungiformis (Gunnerus) Setchell & N.L.Gardner remained while three new species were described. Excluded from Ralfsia sensu stricto were R. pacifica Hollenberg and R. verrucosa (Areschoug) Areschoug. Of the four species of Ralfsia sensu stricto recognized by Parente & Saunders (Citation2019), only R. robertii Parente & G.W.Saunders was confirmed in Europe, although European R. fungiformis-like specimens were not included in that study. Based on traditional taxonomic studies emphasizing morphology (excluding Stragularia spp. and/or scytosiphonacean crustose phases) four species of Ralfsia are currently recognized in European waters: Ralfsia fungiformis reported for Iceland; R. verrucosa widely reported from Iceland, Norway down to the Canary Islands, and also for the Black Sea and Mediterranean; R. ovata Rosenvinge reported for Iceland; R. lucida S.Lund reported for the Baltic Sea (Guiry & Guiry, Citation2018).
The aim of the present study was to investigate species-level taxonomy of Ralfsia-like crusts (excluding Ralfsia sensu stricto (see Parente & Saunders, Citation2019), Stragularia spp. and/or Scytosiphonaceae crustose phases) with an emphasis on the North Atlantic Ocean using COI-5P and rbcL to assist alpha taxonomy and place these species into a phylogenetic context.
Materials and methods
Morphology
Ralfsia-like specimens (n = 75) were collected from many locations with an emphasis on the European coasts of France (Mediterranean coasts, n = 2), Norway (n = 3), Portugal (mainland (n = 11), the Azores (n = 19) and Madeira (n = 3) archipelagos), England (n = 23), with additional collections from North American coasts of Canada (Pacific (n = 7) and Atlantic (n = 5) coasts) and the USA (Atlantic coast, n = 2) (Supplementary table S1). Specimens were dried (pressed or in silica gel) or preserved in 4% formaldehyde solution to serve as vouchers with subsamples preserved in silica gel for molecular analyses (Saunders & McDevit, Citation2012). Specimens collected during the present survey are deposited in the Connell Memorial Herbarium at the University of New Brunswick in Fredericton and in the Herbarium Ruy Telles Palhinha at the University of the Azores in São Miguel. Type specimens of R. confusa Hollenberg, R. integra Hollenberg and R. pacifica were provided on loan from the US National Herbarium (US) and of R. verrucosa from the Swedish Museum of Natural History (S). Sections were made from portions of samples using a freezing microtome (CM 1850, Leica, Heidelberg, Germany) and both sections and squash mounts of thalli were stained with 1% aniline blue in 6% 5 N HCl and mounted in 50% corn syrup with 4% formaldehyde. Observations were made and microphotographs recorded on a Leica DFC480 or Leica CH-9435 digital camera mounted on a Leica DM5000B or a Leica DM2500 light microscope, respectively (Leica, Heidelberg, Germany). Measurements of vegetative and reproductive structures are always presented as height (perpendicular to the substratum) and length/width (parallel to the substratum).
DNA extraction, amplification and sequencing
Total DNA was extracted and the mitochondrial COI-5P (DNA barcode region 5’ end of the mitochondrial cytochrome c oxidase I gene; n = 66; 658 sites) and the nuclear ITS (internal transcribed spacer of the ribosomal cistron, whole region ITS1-5.8S-ITS2; n = 8; 919 sites) were amplified according to Saunders & McDevit (Citation2012) and McDevit & Saunders (Citation2017), respectively. The full fragment of rbcL was also amplified (n = 17; 781 sites), both to assign genetic groups based on rbcL-3P and to perform phylogenetic analyses. For species delineation using rbcL-3P only unidirectional sequence data from the reverse external primer were generated (Saunders & Moore, Citation2013). The primers used are recorded with each GenBank accession (Supplementary tables S1 and S2, BOLD Dataset: RGWS2EUR). For phylogenetic analyses the rbcL was sequenced using the external primers 33F (Draisma et al., Citation2010) and 1381R (Burrowes et al., Citation2003), as well as the internal primers 543F (Bittner et al., Citation2008), PRB-R1A (Kogame et al., Citation1999) and KR2 (Lane et al., Citation2006). PCR products were sequenced using a Big Dye terminator cycle sequencing kit v3.0 (PE Applied Biosystems, Foster City, Canada) and an ABI 3130XL sequencer.
Molecular analyses
Identification of genetic groups was performed through barcode gap analyses (based on raw p-distances) of the COI-5P (n = 66; 658 sites) and rbcL-3P (n = 20; 781 sites) data in BOLD and Geneious 10.2.3 (Kearse et al., Citation2012), respectively. Phylogenetic analyses were performed using each of the two genes separately as well as a concatenated alignment (species and sequences included in the alignments are listed in Supplementary table S1), in all cases incorporating additional COI-5P and rbcL sequence data for other phaeophycean species available in GenBank (Supplementary table S2). To expand our phylogenetic trees rbcL and COI-5P sequences of closely related orders (Fucales, Nemodermatales and Tilopteridales) were retrieved from GenBank (Supplementary tables S2) and trees were rooted with members of Dictyotales (Dictyopteris polypodioides (D.C.) J.V.Lamouroux and Dictyota dichotoma (Hudson) J.V.Lamouroux), Sphacelariales (Cladostephus spongiosus (Hudson) C. Agardh) and Syringodermatales (Syringoderma phinneyi E.C.Henry & D.G.Müller) based on published phylogenies in Silberfeld et al. (Citation2010). Prior to the combined analyses, COI-5P (n = 55; 658 sites) and rbcL (n = 75; 1301 sites) alignments were analysed individually to test for topology congruence. The concatenated alignment of 1959 sites included 75 taxa, all of which had rbcL data and 55 for which COI-5P data were available (Supplementary table S2). Phylogenetic inferences were completed using RAxML (Stamatakis, Citation2014) in Geneious R8 to run a Maximum likelihood (ML) analysis with the model GTR+I+G, partitioned by gene and codon, with 1000 bootstrap replicates.
Results
Identification of species
The DNA barcode analyses of Ralfsia-like crusts (i.e. excluding Ralfsia sensu stricto, Stragularia spp. and/or Scytosiphonaceae crustose phases) revealed 11 strongly divergent genetic groups (only Pseudoralfsia sp._4MP and sp._5MP were closely allied, with 2.27% divergence in their COI-5P sequences; ). Of these 11 genetic groups, only one was assignable to a known species (discussed below), Ralfsia verrucosa (= Pseudoralfsia verrucosa (Areschoug) Parente, Fletcher & G.W.Saunders gen. et comb. nov.). This species had COI-5P intraspecific divergence of 0.33% and was 10.69% divergent from its nearest neighbour Pseudoralfsia sp._10MP, while rbcL-3P divergence was 1.54% from P. sp._3MP (P. sp._10MP lacks COI-5P data) (). Pseudoralfsia verrucosa (n = 27) was found on the coast of Norway (n = 3), the UK (n = 10), and the North American Atlantic (n = 7) and Pacific (n = 7) coasts (Supplementary table S1).
Table 1. Intra- and interspecific variation in COI-5P and rbcL-3P for species of Pseudoralfsiaceae
Pseudoralfsia azorica Parente, Fletcher & G.W.Saunders sp. nov. was based on two samples with a single COI-5P haplotype from the Azores and 19.46% divergence from their nearest neighbour, P. sp._5MP ().
Owing to the close relationship for Pseudoralfsia sp._4MP and P. sp._5MP (2.27% divergence in COI-5P; ), ITS data were also generated, but regrettably for only three specimens – one for P. sp._4MP and two for P. sp._5MP. Only three fixed differences were noted between P. sp._4MP and P. sp._5MP ITS sequences (two substitutions and one indel of five nucleotides), which calls into question two distinct genetic groups, but more study is needed.
Nuchella vesicularis Parente, Fletcher & G.W.Saunders gen. et sp. nov. (n = 11) had COI-5P intraspecific divergence of 0.91% (), ITS intraspecific divergence of 0.27% and 11.55% COI-5P divergence from its nearest-neighbour, N. sp._1MP (n = 1). It was also divergent for rbcL-3P (6.36%) from its nearest neighbour P. verrucosa (). The former species was found on the coasts of mainland Portugal (n = 9), Madeira (n = 1) and the UK (n = 1).
Phylogenetic analyses
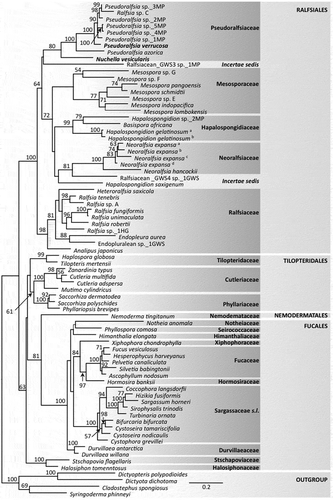
ML analysis of single genes were largely consistent where branches had support and an analysis combining the two regions was completed. The analysis for the concatenated rbcL and COI-5P gene dataset for specimens collected by us and identified as Ralfsia-like resolved three distinct lineages among recognized families of the Ralfsiales – the first proposed here as the new family Pseudoralfsiaceae Parente, Fletcher & G.W.Saunders fam. nov., as well as two distinct lineages among other families of Ralfsiales that will be presented in a future publication (provisionally named Ralfsiacean_GWS3 and GWS4, ). Pseudoralfsia was fully supported in the concatenated alignment and included eight genetic groups, which joined Nuchella in the moderately supported (80% bootstrap support) new family Pseudoralfsiaceae (). In total, Pseudoralfsiaceae included 11 genetic groups (N. sp._1MP and P. sp._10MP lacked rbcL data, and were not included in the concatenated analysis, ) with three of them formally described here. The Pacific Ralfsia sp. C (AB250075, Lim et al., Citation2007) joined the family and resolved within Pseudoralfsia ().
Fig. 1. Maximum likelihood (RAxML) results for the concatenated rbcL and COI-5P alignment. Support values are ML bootstrap values (branches lacking values had < 50% support); the scale bar indicates substitutions per site. Bold type indicates the type specimens of Pseudoralfsia and Nuchella. a, b, c, d are used to distinguish samples of the same species (Supplementary table S2)
Taxonomic results
Pseudoralfsiaceae Parente, Fletcher & G.W.Saunders, fam. nov.
Description
Crusts circular or indefinite in outline, with or without rhizoids and with or without superimposed thalli; erect filaments straight or curved upwards (rarely curved downwards); cells with a single chloroplast lacking obvious pyrenoids; hair pits frequent, arising from the middle and/or lower cells of erect filaments. Unangia sessile, or on 1, rarely 2–3 pedicel cells, on the terminal erect filaments and accompanied by 1–2 paraphyses; sori of plurangia non-adventitious, with synchronized and non-synchronized development of the plurangia; plurangia uniseriate or biseriate, intercalary on the erect filaments with 1, occasionally 2, sterile terminal cells.
Type genus: Pseudoralfsia Parente, Fletcher & G.W.Saunders gen. nov.
Pseudoralfsia Parente, Fletcher & G.W.Saunders, gen. nov.
Description
Thalli circular or indefinite in outline, with or without rhizoids and with or without superimposed thalli; erect filaments straight or curved upwards (rarely curved downwards); cells with a single chloroplast lacking obvious pyrenoids; hair pits frequent arising from the lower cells of erect filaments. Unangia, sessile or on pedicels of 1, rarely 2–3 cells, on terminal erect filaments and accompanied by 1–2 paraphyses; sori of plurangia are non-adventitious with a synchronized development, plurangia uniseriate or at times biseriate, intercalary in the erect filaments with a sterile terminal cell.
Type species: Pseudoralfsia verrucosa (Areschoug) Parente, Fletcher & G.W.Saunders comb. nov.
Etymology: The species of this genus have been confused with true Ralfsia and for that reason we assign the name ‘false Ralfsia’.
Pseudoralfsia verrucosa (Areschoug) Parente, Fletcher & G.W.Saunders comb. nov. ()
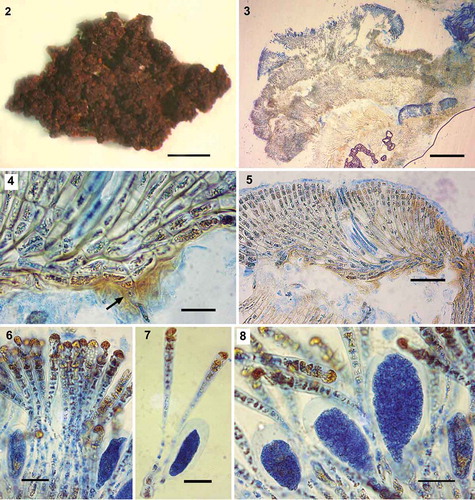
Figs 2–8. Pseudoralfsia verrucosa (Areschoug) Parente, Fletcher & G.W.Saunders comb. nov., observations of the lectotype (Ralfsia verrucosa, A4006). Fig. 2. Image of a portion of the thallus. Fig. 3. Radial-vertical section showing successive small lobes. Fig. 4. Radial-vertical section showing basal cells and one rhizoid (arrow) and erect filaments curving upward. Fig. 5. Radial-vertical section showing one hair pit and erect filaments curving upward. Fig. 6. Unangia and associated paraphyses. Fig. 7. Sessile unangia associated with two paraphyses. Fig. 8. Unangia and associated paraphyses. Scale bars: Fig. 2, 0.3 cm; Fig. 3, 200 µm; Fig. 4, 20 µm Fig. 5, 50 µm; Figs 6–8, 20 µm
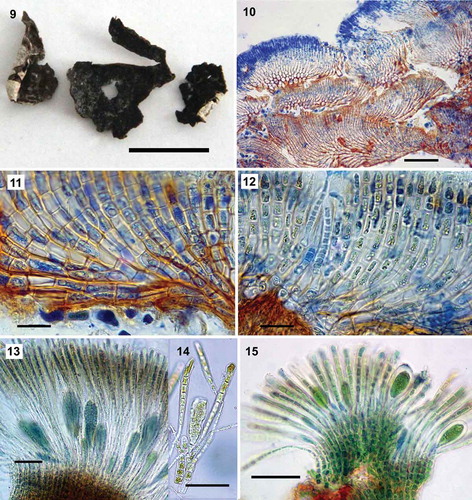
Figs 9–15. Pseudoralfsia verrucosa (Areschoug) Parente, Fletcher & G.W.Saunders comb. nov., based on specimens genetically assigned to this species. Fig. 9. Image of a crust growing in the intertidal excised from rock (MS109). Fig. 10. Radial-vertical section showing successive small lobes (GWS014113). Fig. 11. Radial-vertical section showing basal cells and erect filaments curving upward (GWS014113). Fig. 12. Radial-vertical section showing a hair pit (GWS014106). Fig. 13. Unangia and associated paraphyses (MS109). Fig. 14. Unangium on a three-celled pedicel (arrow) associated with two paraphyses (arrowheads, MS109). Fig. 15. Unangia and associated paraphyses (MS313). Scale bars: Fig. 9, 1 cm; Fig. 10, 100 µm; Figs 11–12, 10 µm; Fig. 13, 60 µm; Figs 14–15, 50 µm
Basionym: Cruoria verrucosa Areschoug, Algarum (phycearum) minus rite cognitarum pugillus secundus. Linnaea. 1843: 264, pl. 9, .
Heterotypic synonyms: Ralfsia verrucosa var. lignicola Areschoug 1847, Ralfsia verrucosa var. cochlearum Areschoug 1876.
Lectotype: Swedish Museum of Natural History, A4006 ().
Type locality: Bohuslän, Western Götaland, Sweden (Areschoug, Citation1843).
Observations on the lectotype (; )
Table 2. Comparison of lectotype to our genetically verified collections assigned to Pseudoralfsia verrucosa (measurements in µm)
Crust dark brown, confluent and indefinite; surface irregular and warty without obvious external concentric zones or radial lines (). Crust typically strongly verrucose as a result of overgrowth of small successive lobes and spaces between lobes () with individual crusts up to 284 µm thick. Underside of crusts rust-red coloured and rarely with rhizoids (); lower stratum mostly composed of horizontally elongated (4–11 µm × 5–32 µm; ), occasionally quadrate (5–6 µm) cells, which give rise to assurgent, rarely descending, curved cell rows. Assurgent curving cell rows grading into erect filaments (), the cells of which were variable in size from quadrate (5–9 µm) to vertically elongated (7–23 µm × 3–11 µm); cells with 1 plate-like chloroplast lacking obvious pyrenoids; hair pits frequent arising from lower cells of erect filaments (). Sori of unangia gelatinous, elevated, frequently in a medium-large sized patch, also in multiple small patches, but never forming continuous sori (). Unangia abundant, of markedly different sizes in the same sorus and between samples (); subclavate or clavate, mainly sessile, but at times on 1, rarely 2, celled pedicels and terminating the erect filaments, accompanied by 1–2 paraphyses. Occasionally immature unangia start to develop early in their maturation process. Mature unangia up to 71 µm high and 37 µm in diameter; paraphyses simple, clavate, up to 13 cells (123 µm) high and up to 12 µm wide at the apex, gradually narrowing with frequently longer cells below ().
Observations of specimens assigned to our genetic group (; )
Crusts light to dark brown, sometimes olive in colour and with a lighter margin, circular in outline or sometimes confluent and indefinite, up to 3 cm broad; crusts typically with an irregular and warty surface without obvious external concentric zones and radial lines (). Crusts strongly verrucose as a result of overgrowth of small successive lobes and spaces between lobes (), or less verrucose or even smooth, with individual crusts up to 270 µm thick. Underside of crusts rust-red coloured; lower stratum mostly composed of horizontally elongated (4–12 µm × 4–35 µm; ), occasionally quadrate (5–12 µm) cells, which give rise to assurgent, rarely descending, curved cell rows. Assurgent curving cell rows grading into erect filaments (), the cells of which were variable in size from quadrate (5–8 µm) to vertically elongated (6–24 µm × 4–12 µm); cells with 1 plate-like chloroplast lacking obvious pyrenoids; hair pits frequent arising from lower cells of erect filaments (). Sori of unangia gelatinous, elevated, frequently in a medium-large sized patch, also in multiple small patches, but never forming continuous sori. Unangia abundant, of markedly different sizes in the same sorus and between specimens (); subclavate or clavate, mainly sessile, but at times on 1, rarely 2–3, celled pedicels and terminating the erect filaments, accompanied by 1–2 paraphyses. Mature unangia 54–115 µm high and 21–38 µm in diameter; paraphyses simple, clavate, 10–14 cells (54–215 µm) high in the central region of mature sori and up to 10 µm wide at the apex, gradually narrowing with frequently longer cells below ().
Habitat, distribution and reproduction: Specimens were collected growing on rock, cobble, stones, molluscs and wood from high upper intertidal to shallow subtidal habitats (to 0.25 m depth), including sheltered and exposed sites, with one sample collected from a salt marsh tidal stream (MS116, GWS022408, GWS030522, GWS022409, GWS007132, MS109, MS427a, MS114, MS313, GWS014106, MS117, MS113, NOR02, MS323, GWS032188, GWS031324, MS112, NOR01, MP11N, GWS014113, GWS008755, GWS005091, GWS027814, NOR03, GWS027815, GWS014465, GWS022407; Supplementary table S1). Specimens were found on European coasts (Norway and UK), as well as north-eastern American coasts (Newfoundland and Labrador, New Brunswick, Nova Scotia and Massachusetts), and north-western American coasts (British Colombia) (Supplementary table S1). Reproductive specimens with plurangia were found in February, May and September (Supplementary table S1).
Pseudoralfsia azorica Parente, Fletcher & G.W.Saunders sp. nov. ()
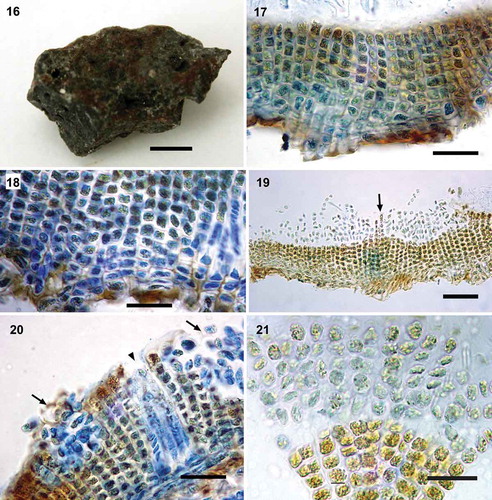
Figs 16–21. Morpho-anatomy of Pseudoralfsia azorica Parente, Fletcher & G.W.Saunders sp. nov. Fig. 16. Image of the holotype (MD0001334c), growing in the upper region of the mid intertidal on rock. Fig. 17. Radial-vertical section showing vertical cell rows (not curved) of erect filaments (MD0001334c). Fig. 18. Radial-vertical section showing basal cells (MD0001334c). Fig. 19. Two slightly elevated sori, separated by two erect filaments (arrow; MD0001334c). Fig. 20. Radial-vertical section showing immature sori separated by erect filaments (arrows) and a hair pit (arrowhead; MD0001334c). Fig. 21. Plurangia with single sterile terminal cells (MD0001334c). Scale bars: Fig. 16, 0.5 cm; Figs 17–18, 20 µm; Fig. 19, 50 µm; Figs 20–21, 20 µm
Description
Crusts brown in colour, lacking a light margin, indefinite in outline (), up to 2 cm broad, smooth, lacking obvious external concentric zones and radial lines. Crusts thin (), up to 220 µm thick; cells of lower stratum mainly horizontally elongate (2.7–5.8 µm × 7.6–14.3 µm; ), which give rise to straight vertical cell rows (). Underside of crusts rust-red and lacking rhizoids; cells of erect filaments ranging from quadrate (4.8–8.1 µm) to vertically elongated (3.6–8.1 µm × 4.1–6.6 µm) and horizontally elongated (3.9–6.3 µm × 4.9–8.8µm); cells with 1 plate-like chloroplast lacking obvious pyrenoids; hair pits abundant arising from lower cells of erect filaments (). Sori gelatinous, non-adventitious and in multiple small-sized patches rather than a single continuous sorus (). Plurangia with up to 8 loci (48 μm high), uniseriate and with 1 sterile terminal cell (6.1–9.6 µm × 4.2–8.8 µm; ); individual loci 4.0–8.8 µm high and 4.3–7.7 µm wide.
Holotype: Specimen MD0001334c (), growing on rock in the upper mid intertidal zone, collected by M. I. Parente and R. Sousa on 8 October 2009 and deposited in the Museu de História Natural de Lisboa.
Type locality: Caloura (37.7, –25.5), São Miguel, Azores, Portugal.
Holotype: DNA barcode: MK972252.
Etymology: Named for its type locality in the Azores.
Habitat, distribution and reproduction: Both specimens were found growing on rock in the upper mid intertidal zone (MD0001334c, MD0001334d; Supplementary table S1). Thus far, only collected in the Azores, São Miguel, Portugal (Supplementary table S1). Reproductive specimen with plurangia was found in October (Supplementary table S1).
Nuchella Parente, Fletcher & G.W.Saunders, gen. nov.
Description
Crusts circular in outline, typically lacking superimposed thalli; sometimes with rhizoids present; erect filaments slightly curving upward (rarely curving downward); cells with a single chloroplast lacking obvious pyrenoids and typically with numerous vesicles; hair pits frequent, arising from both lower and middle cells of the erect filaments. Plurangia embedded in concave, non-adventitious sori with non-synchronous development. Plurangia uniseriate or biseriate, intercalary in erect filaments with 1, occasionally 2, sterile terminal cells.
Type species: Nuchella vesicularis Parente, Fletcher & G.W.Saunders sp. nov.
Etymology: Named in honour of Dr Ana Cristina Costa (affectionately known as Nucha), a valued friend of the senior author (MIP), in recognition of her constant encouragement.
Nuchella vesicularis Parente, Fletcher & G.W.Saunders, sp. nov. ()
Figs 22–27. Morpho-anatomy of Nuchella vesicularis Parente, Fletcher & G.W.Saunders sp. nov. Fig. 22. Image of the holotype (MD0001687), growing in the intertidal on rock. Fig. 23. Radial-vertical section showing slightly curved cell rows (MD0001682). Fig. 24. Radial-vertical section showing multicellular rhizoids (MD0001681). Fig. 25. Upper cells of erect filaments filled with many intracellular vesicles (MD0001687). Fig. 26. Radial-vertical section showing a non-adventitious sorus (arrow; MD0001687). Fig. 27. Detail of a sorus with uniseriate and biseriate plurangia (arrows) capped by one or two sterile terminal cells (arrowheads; MD0001687). Scale bars: Fig. 22, 0.5 cm; Fig. 23, 100 µm; Fig. 24, 50 µm; Fig. 25, 20 µm; Fig. 26, 50 µm; Fig. 27, 20 µm
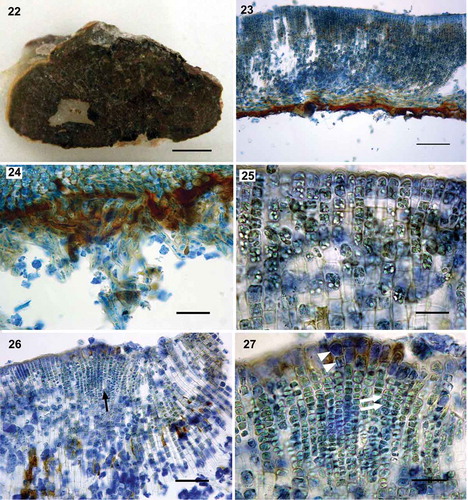
Description
Crusts light to medium brown in colour sometimes with a lighter margin, circular in outline (), up to 3 cm broad; surface regular and smooth, typically with radial lines and sometimes with concentric zones. Typically lacking superimposed thalli; 120–417 µm in thickness (); underside of crusts rust-red coloured, occasionally with multicellular rhizoids (). Lower region of stratum composed mostly of 4 to 6 layers of horizontally elongated (2.4–15.5 µm × 9.1–27.5 µm) and occasionally quadrate (3.7–15.5 µm) cells, which give rise to assurgent (), rarely descending, slightly curving cell rows; middle cells of erect filaments of variable sizes from small quadrate (7–10 µm), often vertically elongated (6.4–25.6 µm × 3–14.8 µm) and at times horizontally elongated (6–15 µm × 10–17 µm); cells of the apical region of the erect filaments often horizontally elongated (4.2–10.6 µm × 5.0–11.6 µm), quadrate (5–11.6 µm) or, also vertically elongated (5.6–10.6 µm × 5–8.6 µm); cells with one plate-like chloroplast lacking obvious pyrenoids and typically with several intracellular vesicles (); hair pits frequent arising from both lower and middle cells of the erect filaments. Sori gelatinous, in multiple, small-sized patches rather than in a single continuous sorus, embedded, concave and non-adventitious with non-synchronous development of the plurangia (); plurangia up to 14 loci, uniseriate, sometimes biseriate with 1 or 2 sterile terminal cells (5.0–16.0 µm × 2.5–5.8 µm; ); individual loci 2.1–7.2 µm high and 2.5–5.7 µm wide; unangia unknown.
Holotype: Specimen MD0001687 (), growing on rock in the mid intertidal zone, collected by M.I. Parente and R. Sousa on 24 July 2010 and deposited in Museu de História Natural de Lisboa.
Type locality: Gaia, São Felix da Marinha, Portugal.
Holotype DNA barcode: MK972260.
Etymology: Named on the basis of the presence of several vesicles in each cell.
Habitat, distribution and reproduction: Specimens were collected growing on rock in the mid intertidal zone in mainland Portugal, Madeira and the UK (MP13N, MD0001675, MD0001660, PG071357, MD0001681, MD0001657, MD0001687, MD0001682, MPFL241; MD0001684, MPFL243, MP31N; Supplementary table S1). Reproductive specimen with plurangia was found in July (Supplementary table S1).
Discussion
The present study, which emphasized North Atlantic and notably European collections, revealed hidden diversity in Ralfsia-like samples. Based on molecular analyses and morpho-anatomical comparisons to type material, we propose to transfer a monophyletic subset of Ralfsia-like species (except Ralfsiacean_GWS3 sp._1MP and Ralfsiacean_GWS4 sp._1GWS unresolved among the Ralfsiales complex of families; ) to the moderately supported new family Pseudoralfsiaceae, Ralfsiales. This family is assigned two genera, Nuchella and Pseudoralfsia, that do not resolve within the currently recognized families (viz. Hapalospongidiaceae, Mesosporaceae, Neoralfsiaceae and Ralfsiaceae; ). Species of Pseudoralfsiaceae are morphologically similar to species of Ralfsia sensu stricto, Ralfsiaceae, but can be distinguished by having frequent hair pits, typically unsymmetrical thalli and, in particular, by their molecular data. Indeed, the deep genetic differences support the recognition of separate families (see ). León-Alvarez et al. (Citation2017) similarly considered sequence data of primary importance in distinguishing the Hapalospongideaceae from the Mesosporaceae, which lack robust distinguishing morphological or anatomical features. Similar decisions were implemented by Hind et al. (Citation2016) regarding the subfamilies Corallinoideae and Neogoniolithoideae and, more recently, by Caragnano et al. (Citation2018) regarding the subfamily Chamberlainoideae, Corallinales. The data presented here support placement of P. verrucosa, P. azorica, N. vesicularis and eight additional informal genetic groups within Pseudoralfsiaceae.
The genetic group that we assigned to Pseudoralfsia verrucosa was distinct from all of the other genetic groups studied here with vegetative construction common to that in the lectotype of R. verrucosa. In both, there is a build-up of several small successive lobes resulting in thick verrucose and convoluted crusts typically lacking a symmetrical shape (see ). This development distinguishes P. verrucosa from all the other Ralfsiales described to date, as well as all of the genetically identified taxa uncovered in the current manuscript. We are thus confident that this genetic group is correctly assigned to Ralfsia verrucosa. Although the lectotype of R. verrucosa and our samples varied with respect to the maximum dimensions of their mature unangia (71 µm × 37 µm and 54–115 µm × 21–38 µm, respectively; ) and maximum length of their paraphyses (123 µm and 54– 15 µm, respectively), the size of paraphyses and unangia of the lectotype are within the size range of our samples. Our collections of P. verrucosa have a widespread distribution (Pacific and Atlantic North American coasts, UK and Norway) including one specimen in close geographic proximity (less than 100 km distant from Verdens Ende, Vestfold, Skagerrak coast of Norway) to the type locality (Bohuslän, Western Götaland, Sweden). This lends support to our morpho-anatomical observations in assigning our genetic group to this morphospecies.
Based on the present results and Parente & Saunders (Citation2019) regarding identification conflicts within crustose species of the Ralfsiales, and the morphological variation displayed among specimens of R. verrucosa (discussed above), all previous reports of R. verrucosa should be confirmed by sequence data.
Pseudoralfsia azorica displayed considerable COI-5P interspecific divergence from other species (19.46%) consistent or greater than that between species and genera of the Ralfsiaceae, as well as between other phaeophycean species and genera (e.g. Lim et al., Citation2007; McDevit & Saunders, Citation2009; Poong et al., Citation2014; León-Alvarez et al., Citation2017; Parente & Saunders, Citation2019). It differs morpho-anatomically from P. verrucosa by having thinner thalli, straight erect filaments (not curving) and in having smaller cell sizes (). Pseudoralfsia azorica differs morpho-anatomically from Ralfsia tenebris Parente & G.W.Saunders in bearing frequent hairs often arising from lower cells of erect filaments, in having much thinner thalli, brown colour, and different habitat preferences (found in the middle or upper littoral on rock surfaces). It also differs from Ralfsia confusa Hollenberg in its brown colour and sori in multiple small patches. The holotype of R. confusa is a light brown crust, mostly with a single continuous sorus situated in the central part of the crust. As P. azorica was based on only two collections (Supplementary table S1), we eagerly await additional information regarding its morphological variation, seasonality, ecological and geographic ranges.
Nuchella was proposed to accommodate the generitype N. vesicularis sp. nov. and an informal genetic group provisionally named Nuchella sp._1MP, which was represented by a single sterile specimen. Nuchella vesicularis is both genetically distinct () and morpho-anatomically different from Pseudoralfsia spp. and other crustose Ralfsiales in having embedded and concave, non-adventitious plurangia sori with non-synchronous development, as well as consistently having numerous vesicles in the cells. Nuchella is tentatively included in Pseudoralfsiaceae until its evolutionary affiliation can be better resolved ().
Author contributions
M.I. Parente: original concept, drafting and editing manuscript, sample descriptions; molecular data analyses and interpretation; R.L. Fletcher: original concept, sample descriptions, editing manuscript; F.O. Costa: original concept, molecular data analyses, editing manuscript; G.W. Saunders: original concept, drafting, editing manuscript, molecular data analyses, sample descriptions.
TEJP-2019-0077-File008.docx
Download MS Word (39.3 KB)TEJP-2019-0077-File007.docx
Download MS Word (48.5 KB)Acknowledgments
Many thanks to previous and current members of the Saunders Laboratory for technical assistance and for valued advice and helpful discussions. We thank all the collectors listed in Supplementary table S1 for their critical involvement in this project. Thanks also to Ana Ferreira, Elisabete Dias, Ricardo Camarinho, Maximilian Müller and Sophia Griese for laboratory help, Ana C. Costa, Mónica Moura and Armindo Rodrigues for making available laboratory equipment and Ana C. Costa also for helpful comments. Thanks are also due to Alvin K. Chan, K. Dixon, D. McDevit and T. Moore for generating some of the sequence data. We gratefully acknowledge the trustees of the US National Herbarium and Swedish Museum of Natural History for organizing loans of type material and providing helpful taxonomic information. We also thank Michael Wynne for assistance in interpreting historical taxonomic literature and Craig Schneider and António Frias Martins for assistance with Latin.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary information
The following supplementary material is accessible via the Supplementary Content tab on the article’s online page at http://dx.doi.org/10.1080/09670262.2020.1753245
Supplementary table S1. Specimens included in the present study with collection information and GenBank accession numbers. Reproductive specimens are indicated with a and b (for plurangia and unangia respectively), and accession numbers of the sequences used in the concatenated alignment are marked in bold type.
Supplementary table S2. Sequences retrieved from GenBank with collection information, publication and accession numbers. a, b, c, d are used to distinguish samples of the same species.
Additional information
Funding
References
- Areschoug, J.E. (1843). Algarum (phycearum) minus rite cognitarum pugillus secundus. Linnaea, 17: 257–269, pl. IX.
- Bittner, L., Payri, C.E., Couloux, A., Cruaud, C., Reviers, B. de. & Rousseau, F. (2008). Molecular phylogeny of the Dictyotales and their position within the brown algae, based on nuclear, plastidial and mitochondrial sequence data. Molecular Phylogenetics and Evolution, 49: 211–226.
- Burrowes, R., Rousseau, F., Müller, D.G. & Reviers, B. de (2003). Taxonomic placement of Microzonia (Phaeophyceae) in Syringodermatales based on rbcL and 28S nrDNA sequences. Cryptogamie Algologie, 24: 63–73.
- Caragnano, A., Foetisch, A., Maneveldt, G.W., Millet, L., Liu, L.-C., Lin, S.-M., Rodondi, G. & Payri, C.E. (2018). Revision of Corallinaceae (Corallinales, Rhodophyta): recognizing Dawsoniolithon gen. nov., Parvicellularium gen. nov. and Chamberlainoideae subfam. nov. containing Chamberlainium gen. nov. and Pneophyllum. Journal of Phycology, 54: 391–409.
- Draisma, S.G.A., Prud’homme Van Reine, W.F. & Kawai, H. (2010). A revised classification of the Sphacelariales (Phaeophyceae) inferred from a psbC and rbcL based phylogeny. European Journal of Phycology, 45: 308–326.
- Guiry, M.D. & Guiry, G.M. (2018). Algaebase. World-wide electronic publication, National University of Ireland, Galway (Online). Available from http://www.algaebase.org ( accessed 18 May 2018).
- Hind, K.R., Gabrielsen, P.W., Jensen, C.P. & Martone, P.T. (2016). Crusticorallina gen. nov., a nongeniculate genus in the subfamily Corallinoideae (Corallinales, Rhodophyta). Journal of Phycology, 52: 929–941.
- Kain, J.M., Buchanan, J., Boo, S.M. & Lee, K.M. (2010). Colpomenia bullosa crust masquerading as Ralfsia verrucosa (Phaeophyceae) in southeast Australia. Phycologia, 49: 617–627.
- Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P. & Drummond, A. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647–1649.
- Kogame, K., Horiguchi, T. & Masuda, M. (1999). Phylogeny of the order Scytosiphonales (Phaeophyceae) based on DNA sequences of rbcL, partial rbcS, and partial LSU nrDNA. Phycologia, 38: 496–502.
- Lane, C.E., Mayes, C., Druehl, L.D. & Saunders, G.W. (2006). A multi-gene molecular investigation of the kelp (Laminariales, Phaeophyceae) supports substantial taxonomic re-organization. Journal of Phycology, 42: 493–512.
- León-Alvarez, D., Reyes-Gómez, V.P., Wynne, M.J., Ponce-Márquez, M.E. & Quiróz-González, N. (2017). Morphological and molecular characterization of Hapalospongidion gelatinosum, Hapalospongidiaceae fam. nov. (Ralfsiales, Phaeophyceae) from Mexico. Botanica Marina, 60: 567–581.
- Lim, P.-E., Sakaguchi, M., Hanyuda, T., Kogame, K., Phang, S.-M. & Kawai, H. (2007). Molecular phylogeny of crustose brown algae (Ralfsiales, Phaeophyceae) inferred from rbcL sequences resulting in the proposal for Neoralfsiaceae fam. nov. Phycologia, 46: 456–466.
- McDevit, D.C. & Saunders, G.W. (2009). On the utility of DNA barcoding for species differentiation among brown macroalgae (Phaeophyceae) including a novel extraction protocol. Phycological Research, 57: 131–141.
- McDevit, D.C. & Saunders, G.W. (2017). A molecular investigation of Canadian Scytosiphonaceae (Phaeophyceae) including descriptions of Planosiphon gen. nov. and Scytosiphon promiscuus sp. nov. Botany, 95: 653–671.
- Nakamura, Y. (1972). A proposal on the classification of the Phaeophyta. In Contributions to the Systematics of Benthic Marine Algae of the North Pacific (Abbott, I.A. & Kurogi, M., editors), 147–155. Japanese Society of Phycology, Kobe.
- Parente, M.I. & Saunders, G.W. (2019). A molecular survey of Ralfsia sensu stricto (Ralfsiales, Phaeophyceae) in Canada uncovers three new species: R. robertii sp. nov., R. tenebris sp. nov. and R. unimaculata sp. nov. Botany. doi.org/10.1139/cjb-2018-0138.
- Poong, S.W., Lim, P.E., Phang, S.M., Gerung, G.S. & Kawai, H. (2013). Mesospora elongata sp. nov. (Ralfsiales, Phaeophyceae), a new crustose brown algal species from the Indo-Pacific region. Phycologia, 52: 74–81.
- Poong, S.W., Lim, P.E., Phang, S.M., Sunarpi, H., West, J.A. & Kawai, H. (2014). A molecular-assisted floristic survey of crustose brown algae (Phaeophyceae) from Malaysia and Lombok Island, Indonesia based on rbcL and partial cox1 genes. Journal of Applied Phycology, 26: 1231–1242.
- Poong, S.W., Lim, P.E., Phang, S.M., Sunarpi, H., West, J.A., Miller, K.A., Nelson, W. & Kawai, H. (2017). Two new species of Mesospora (Ralfsiales, Phaeophyceae) from the subtropical Indo-Pacific region. Phycologia, 56: 487–498.
- Saunders, G.W. & McDevit, D.C. (2012). Methods for DNA barcoding photosynthetic protists emphasizing the macroalgae and diatoms. Methods in Molecular Biology, 858: 207–222.
- Saunders, G.W. & Moore, T.E. (2013). Refinements for the amplification and sequencing of red algal DNA barcode and RedToL phylogenetic markers: a summary of current primers, profiles and strategies. Algae, 28: 31–43.
- Silberfeld, T., Leigh, J.W., Verbruggen, H., Cruaud, C., De Reviers, B. & Rousseau, F. (2010). A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): investigating the evolutionary nature of the “brown algal crown radiation”. Molecular Phylogenetics and Evolution, 56: 659–674.
- Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analyses and post-analysis of large phylogenies. Bioinformatics, open access.
