Abstract
Background: Genomic aberrations are important indicators of prognosis, clinical course and treatment of chronic lymphocytic leukemia (CLL). Two cytogenetic methods, karyotype, and FISH, with still ongoing improvements, are used for CLL investigation, but the panel of chromosomal abnormalities, their prognostic significance and contribution in CLL pathogenesis have not been elucidated yet.
Objectives and methods: Our study deals with the cytogenetic investigation of 237 CLL patients trying to answer ambiguous issues of the disease in the light of new CLL stimulation methodology. More specifically, we compared the detection rate and type of chromosomal aberrations between cultures stimulated with and without the new mitogens and we combined them with the data obtained from interphase (iFISH) and metaphase FISH (mFISH).
Results: Approximately 70% of the abnormal karyotypes and all the subclonal abnormalities were detected exclusively in DSP-30/IL-2 cultures. DSP-30/IL-2 exhibited ∼10-fold greater ability to detect abnormalities compared to TPA and unstimulated cultures, revealing >60 different chromosomal aberrations. Moreover, the comparison between DSP-30/IL-2 cultures and unstimulated cultures indicated that loss of chromosome Y is rather an age-related phenomenon and not a specific aberration of CLL. Clonal evolution was also detected in 50% of patients with available follow-up karyotypic data and changed the prognosis in 86.4% of them. Finally, it was shown that mFISH must be performed in DSP-30/IL-2 cultures in addition to iFISH to uncover submicroscopic translocations or insertions undetectable by iFISH.
Conclusion: All the above argue in favor of the parallel application of karyotype, iFISH and mFISH after DSP-30/IL-2 stimulation for CLL clinical practice and research.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by a remarkably heterogeneous clinical course ranging from very indolent cases to patients with aggressive and rapidly progressing disease. This heterogeneity has important consequences on clinical approaches, treatment strategies and survival.Citation1
A significant prognostic factor correlating with clinical course, treatment and follow-up of CLL is the presence of chromosomal aberrations.Citation2 Two cytogenetic techniques with still ongoing improvements, conventional chromosome G-banding analysis (CBA) and fluorescence in situ hybridization (FISH) are used as standard methods to detect genomic aberrations in CLL.Citation3,Citation4 CBA provides a whole genome perspective, but its diagnostic yield is hampered by the low mitotic activity of CLL cells in vitro. Recently, the introduction of the new CLL stimulation methodology with CpG-oligonucleotides plus interleukin-2 (IL-2) has been reported to enhance the proliferation of B-CLL lymphocytes.Citation5,Citation6 On the other hand, FISH allows the detection of submicroscopic chromosomal aberrations which are undetectable by CBA, irrespectively the cells ability to divide. However, the information gained by this method is limited to the targets of the selected probes. Consequently, the panel of chromosome abnormalities and their prognostic significance in CLL have not been fully determined yet while the prognostic importance of less frequent or rare recurrent aberrations is controversial, or still unknown.
Clonal evolution (CE) with the acquisition of new cytogenetic aberrations may occur during the course of the disease and sometimes may be associated with shorter overall survival.Citation7 However, the types of chromosomal lesions which occur during the course of the disease and their prognostic significance have not been elucidated yet. CE detected by CBA and FISH could be proved as important indicators during the clinical course of the disease.
In the present study, we tried to deal with important issues of the cytogenetic investigation of CLL in the light of new CLL stimulation methodology. More specifically, we compared the detection rate and type of chromosomal aberrations between cultures stimulated with and without the use of new mitogens and we combined them with the data obtained from interphase FISH (iFISH) and metaphase FISH (mFISH) in order to point out the best methodology for the cytogenetic analysis of CLL. We also tried to elucidate whether loss of chromosome Y is an abnormality related to CLL or an age-related phenomenon. Finally, using the new mitogens, we assessed the frequency of CE and the types of acquired chromosomal abnormalities during the course of the disease.
Patients and methods
Patients
Study population was consisted of 237 CLL patients. CLL was diagnosed by experts according to IWCLL criteria, between 2008 and 2012 in Greece.Citation1 Bone marrow samples obtained at the time of diagnosis and follow-up of the patients were sent to our laboratory for cytogenetic analysis. All samples were processed with written informed consent in accordance with the Declaration of Helsinki under protocols approved by the local ethical committee.
Chromosome G-banding analysis
Bone marrow samples (400 μL) were cultivated in complete McCoy's 5Α (Biochrom) medium with 10% fetal bovine serum. Three types of cultures were set up in parallel for all samples: (a) a 24 hours and a 48 hours unstimulated culture (b) a 72 hours stimulated with 12-O-tetradecanoylphorbol-13-acetate (TPA) (20 ng/mL; Sigma-Aldrich) culture and (c) a 72 hours stimulated with CpG-oligonucleotide DSP-30 (2 Μμ; TIB Mol-biol, Berlin, Germany) and IL-2 (200 U/mL; Roche Diagnostics, Germany) culture. Colcemide (0.02 μg/mL, Sigma, St. Louis, MO, USA) was added to the culture for the last hour of incubation. Slide preparations were performed according to standard cytogenetic protocols. Imaging and karyotyping were performed via microscopy and computer imaging techniques. Thirty metaphases per sample, 10 of each type of culture (if possible) were karyotyped. Karyotypic analysis was considered successful when an abnormal clone was detected or ≥20 normal metaphases were analyzed per sample. Karyotypes were analyzed according to the International System for Human Cytogenetic Nomenclature (ISCN) 2013. The abnormality detection rate was defined as the proportion of abnormal cases detected from a particular method or type of culture against the total number of successfully analyzed CLL cases. Complex karyotype was defined as a karyotype with ≥3 clonal aberrations.
Fluorescence in situ hybridization
FISH was carried out on stimulated with DSP-30 plus IL-2 (DSP-30/IL-2) cytogenetic specimens of 126 out of 237 CLL patients, due to the high cost of FISH analysis. We chose DSP-30/IL-2 culture in order to be able to analyze not only interphase cells but also metaphases of CLL cells. The commercial CLL set probes were used for detection of the most common abnormalities of the disease including deletions of 17p13 (TP53), 11q22.3 (ATM), and 13q14.3/13q34.3 (D13S319/13q34) regions and trisomy 12 (CEP 12) (Vysis Inc., Downers Grove, IL). Occasionally, other FISH probes were also used for further investigation and/or confirmation of specific abnormalities found by karyotype. All DNA probes were applied following standard procedures outlined by the manufacturers. At least a total of 200 interphases and 20 metaphases were evaluated in each patient. Dual-color FISH images were digitally generated using Isis FISH-imaging software (MetaSystems, Altlussheim, Germany).
Statistical analysis
The statistical significance of differences in study subgroups of CLL patients was calculated using χ2-statistics with continuity correction or Fisher's exact test. P-value less than 0.05 was considered statistically significant. Odds ratio (OR) was calculated and reported with 95% confidence intervals (95% CI).
Results
Demographic characteristics
Patients’ baseline characteristics are presented in Table . The sex ratio (males/females) was 1.63. Patients’ mean age was 64.9 years, with no significant differences between the two sexes.
Table 1 Demographic characteristics of CLL cases
Comparison of CLL aberrations between stimulated and unstimulated cultures
Aberrant clones where detected in (a) DSP-30/IL-2 cultures uniquely (43.1%), (b) cultures with no mitogens uniquely (3.7%), (c) both DSP-30/IL-2 and unstimulated cultures (3.7%), (d) both DSP-30/IL-2 and TPA cultures (6.0%), and (e) all cultures (8.3%). The identification of the best stimulator for CLL cells required the comparison of the detection rate and type of clonal aberrations among the three types of cultures which is observed in Table .
Table 2 Abnormality detection rate among cultures with DSP/IL-2, TPA and no mitogens
More specifically the new combination of mitogens DSP-30/IL-2 revealed clonal aberrations in 58.7% of the successfully analyzed CLL cases while unstimulated cultures and stimulated with TPA-revealed clonal aberrations in 12.4% each (Table ). Interestingly, there were 128 cases (∼70% of the abnormal karyotypes) with the abnormal clones detected solely in DSP-30/IL-2 cultures while a normal karyotype was observed in their corresponding TPA and unstimulated cultures. No clonal aberration was revealed solely in TPA cultures.
On the other hand, there were eight cases (5.9%) that revealed aberrations only in unstimulated cultures. These patients demonstrated loss of chromosome Y (−Y) as a sole aberration. Beyond this, all the clonal abnormalities detected in unstimulated or stimulated with TPA cultures were also observed in DSP-30/IL-2 cultures. Comparing the abnormality rate of cultured metaphase cells, the increase in the detection rate for DSP-30/IL-2 over the other two culture was statistically significant (P < 0.0001). In particular the combination of DSP-30/IL-2 exhibited approximately ∼10-fold greater ability to detect the abnormal clone compared to TPA and unstimulated cultures (OR = 10.06, 95% CI = 6.2–16.3, P > 0.0001). In addition, DSP-30/IL-2 also yielded the highest overall percentage of abnormal cells while all the existing subclonal abnormalities were detected only in DSP-30/IL-2 cultures.
CBA
Karyotypic analysis was successful in 218 patients (92.0%). Normal karyotypes were found in 82 (37.6%) and abnormal in 136 patients (62.4%). No significant differences were observed between the two sexes regarding the distribution of normal and abnormal karyotypes. Among the abnormal karyotypes, 65 (47.8%) presented a single aberration while 43 (31.62%) were complex. The analysis of the abnormal karyotypes revealed more than 60 different recurrent common or rare chromosomal aberrations. The most common chromosomal aberration was trisomy 12 (+12) (21.10% of the total successfully analyzed CLL cases) followed by del(13q) (10.55%), translocations of 14q (9.6%), −Y (7.3%), del(11q) (6.0%), del(6q) (5.5%), and −17 (4.6%) (Table ). On the other hand, del(17p) which confers the worst prognosis in CLL, was detected in only three patients (1.4%) by karyotype, as it is usually a submicroscopic abnormality. Nevertheless, −17, resulting in del(17p), was revealed in ∼5% of the patients and only in complex karyotypes. Other monosomies such as −5, −8, −18, −14, and −X which were also found only in complex karyotypes are not considered as CLL specific aberrations. Moreover, in our sample, chromosome losses (38.53%) and deletions of chromosomal regions (33.03%) were prevailing over trisomies (30.73%).
Table 3 The most common karyotypic abnormalities and their frequencies in CLL patients
Loss of chromosome Y was the second commonest aberration as a sole abnormality. The mean age of these patients was 72.4 years, older than that of the general CLL group found in the study (64.9 years). Karyotypic data of patients with −Y are presented in Table . As demonstrated, −Y was almost always present in the unstimulated cultures of the patients irrespectively the presence of additional aberrant clones in the stimulated cultures. Moreover, 56.3% of patients with –Y, demonstrated the loss only in their unstimulated cultures while a normal karyotype was demonstrated in DSP-30/IL-2 cultures. When –Y was found in both stimulated and unstimulated cultures, the percentage of the aberrant clone was prevailing in the latter case. All the above indicate that −Y is rather a consequence of the advanced age and not a CLL specific abnormality.
Table 4 Cytogenetic data of patients with loss of chromosome Y
Furthermore, a variety of rare chromosomal aberrations, defined as those found in at least two patients (either in this study or in the data base of Mitelman) so as to be recurrent and in less than 2% of CLL cases, was mainly observed in DSP-30/IL-2 cultures. These abnormalities are shown in Table . Trisomies and monosomies were detected mainly in complex karyotypes although +7 and +19 were also met as sole aberrations in our CLL group. The chromosomal regions 2q27, 7q32, 16q22–24, and 19p13 were implicated in abnormal karyotypes as derivative chromosomes.
Table 5 Rare or novel recurrent chromosomal aberrations found in our CLL group
Clonal evolution
Diagnostic and follow-up karyotypic data (median 31.7 months, range 4–73 months) were available in 44 patients. Clonal devolution, defined as the disappearance of one or more clonal aberrations at follow-up, was identified in 15.9% (7/44) patients. CE was detected in 50% (22/44) of the patients (Table ). The most common newly detected aberrations during the course of the disease were +12 (27.3%), −Y (27.3%), and del(13q) (22.7%) and abnormalities of chromosomes 18 (22.7%) and 14 (22.7%). The new cytogenetic abnormalities changed the prognostic classification of 19 out of 22 patients (86.4%); 11 of them presented new aberrations with worse prognosis while the new aberrations in three patients conferred better prognosis. More specifically 11 patients with a normal karyotype at diagnosis demonstrated newly acquired aberrations such as +12 (three patients), del(13q) (three patients), −Y (three patients), t(14;18)(q32;q21) (one patient), −17 (one patient) and complex karyotype (six patients). Peculiarly, we detected at baseline four patients with inv(9)(p11q13), which is generally considered a normal variation, as a sole aberration, who acquired more than three additional aberrations, including del(13q), resulting in complex karyotypes, during the course of the disease.
Table 6 Clonal evolution
iFISH and mFISH
iFISH analysis was successful in all 126 patients while aberrations were detected in 87 patients (69.05%). In detail, del(13)(q14.3) was found in 57 patients (45.24%), +12 in 23 (18.25%), del(17)(p13.1)/p53 in 13 (10.32%), del(11)(q22.3)/ATM in 11 (8.73%), and del(13)(q34.3) in 2 (1.59%) patients. Moreover, a homozygous deletion of 13q14.3 was found in 13 (10.32%) patients. No significant differences were noticed between the gender and the total number of FISH aberrations or a specific abnormality.
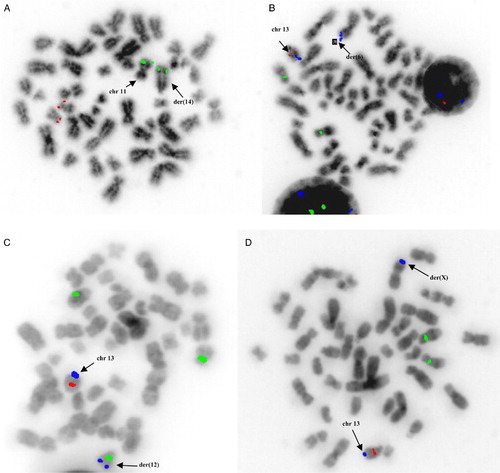
FISH was performed on stimulated with DSP30/IL-2 cells so as to be able to analyze not only interphase but also metaphase CLL cells. mFISH detected unexpected abnormal hybridization patterns compared to those obtained by iFISH using the same CLL set probes. More specifically, we noticed the following translocations, undetectable by iFISH, in four patients: the chromosomal region 11q22.3 (ATM) was translocated on 14q (Fig A), while 13q34.3 was found to be translocated on 6p (Fig B), 12q (Fig C) and the short arm of chromosome X (Fig D) (one translocation in each patient).
Figure 1. Translocations undetectable by iFISH was observed in four patients by mFISH: (a) the chromosomal region 11q22.3 (ATM) (green signal) was translocated intact on 14q (red signal corresponds to 17p13.1/p53), (b) the chromosomal region 13q34.3 (blue signal) was found to be translocated intact on 6p (green signal corresponds to CEP12 and red to 13q14.3), (c) the chromosomal region 13q34.3 (blue signal) was found to be translocated intact on 12q (green signal corresponds to CEP12 and red to 13q14.3), and (d) the chromosomal region 13q34.3 (blue signal) was translocated intact on the short arm of chromosome X (green signal corresponds to CEP12 and red to 13q14.3).
CBA vs. FISH
Sixty-five of the 126 cases (51.6%) who were analyzed by FISH had normal or no karyotypic data. Among these cases, FISH revealed submicroscopic deletions in 53.85% (35/65), not visible by karyotypic analysis. More specifically 30 patients demonstrated del(13)(q14.3), one patient del(11)(q22.3) and four patients del(17)(p13.1) which confers an adverse prognosis.
On the other hand, among 61 cases with abnormal karyotype and available FISH data, the karyotypic results of 46 patients (75.4%) provided additional information to FISH while in 18 of them karyotype altered the prognosis of FISH results (Table ). These patients presented mostly complex karyotypes including translocations and other rearrangements of chromosomal regions undetectable by the current CLL FISH panel. Thus, the poor prognosis of the karyotypic aberrations altered the good prognosis of FISH abnormalities. Moreover, cases with del(11)(q22.3)/ATM and del(17)(p13.1)/p53 detected by FISH, showed complex karyotypes in 60.0% and 54.0%, respectively.
Table 7 Patients with karyotypes which altered the prognosis of FISH results
Discussion
Recently, synthetic CpG-oligodeoxynucleotides (CpG-ODN) have been used to stimulate cell division in CLL specimens and enhance the detection rate of clonal aberrations.Citation8,Citation9 These oligonucleotides induce proliferation, cytokine production, and high-affinity IL-2 receptor expression in CLL cells through interaction with Toll-like receptor 9 on B cells.Citation10,Citation11 Prior research studies showed that IL-2 activated by CpG-ODN prevents death of CLL cells and thus the combination of IL-2 with CpG-ODN can effectively induce cell cycle progression of CLL cells in vitro enabling the detection of clonal aberrations.Citation12
DSP-30/IL-2 enhances the detection rate of abnormal clones compared to TPA and unstimulated cultures
Our study which is the largest one using three different types of cultures for the estimation and comparison of detection rate and type of clonal aberrations demonstrated that DSP-30/IL-2 significantly enhances the detection rate of abnormal clones (approximately 10-fold) compared to TPA and unstimulated cultures. Notably, ∼70% of the abnormal cases were detected solely in DSP-30/IL-2 cultures while normal karyotypes were observed in their corresponding TPA and unstimulated cultures. By means of DSP-30/IL-2 stimulation methodology we detected abnormal karyotypes in 62.4% of the patients, overcoming the line of 30–50% abnormal karyotypes due to the traditionally used mitogens.Citation13 Therefore, for clinical practice, we suggest that priority should be given to DSP-30/IL-2 cultures and then to TPA or unstimulated cultures.
Karyotypic analysis enlightened controversial issues of the disease
In the current study, karyotypic analysis enlightened controversial issues of the disease including the yield of chromosomal aberrations in CLL, the nature of –Y and the chromosomal aberrations implicated in CE.
In particular, whether loss of chromosome Y is an age-related phenomenon or a cytogenetic marker of hematological disease is still unclear. The fact that in our study, –Y was found in older patients and often as a single aberration only in unstimulated cultures while the corresponding stimulated cultures with DSP-30/IL-2 were normal indicates that –Y is not a CLL specific aberration but rather a consequence of the advanced age. The detection of –Y in both unstimulated and stimulated cultures but accompanied by other CLL abnormalities only in stimulated with DSP-30/IL-2 cultures may be possibly attributed to the transformation of a normal cell with –Y (as a consequence of the advanced age) into a CLL cell after the acquisition of specific CLL chromosomal aberrations (especially del(13q) and/or +12). In the latter case, loss of chromosome Y could provide a proliferative advantage simply because it tends to replicate late in S-phase and its loss might shorten slightly the cell cycle.Citation14 This finding is not in accordance with the only study investigating sex chromosome loss in CLL which approached this issue by a different method and considered –Y as a CLL specific aberration.Citation15
Due to the ability of karyotypic analysis to uncover chromosomal abnormalities through the whole genome, we were also able to detect a variety of chromosomal abnormalities including the rare ones, contributing to the identification of the whole panel of CLL cytogenetic abnormalities. Among the novel aberrations demonstrated in our CLL group was t(6;13)(p21;q14) as a secondary event to the interstitial deletion of 13q14.3 region. We have already reported that this translocation is resistant to the current treatment CLL protocols but without conferring an adverse prognosis.Citation16 Deletions of chromosome 5 which were also detected in our patients are rare abnormalities in CLL.Citation17 Moreover der(4)t(4;17) accompanied by −17 was detected for the first time in two of our patients and thus it could be characterized as a recurrent rare abnormality, accompanied by −17. Another rare translocation, t(2;13)(q21–23;q12–34), already reported, was found in three patients.Citation18 Chromosomal abnormalities implicating regions 2p, 3q, and 7q which were observed in our patients, have been recently associated with adverse outcome.Citation19
CE was detected in 50% of our patients with available follow-up data by CBA, while a recent study using FISH analysis detected CE in 14.3% of the patients examined, indicating that CBA is a better method than FISH for this investigation.Citation20 In our study, the most common newly detected aberrations during the course of the disease were +12, −Y, del(13q) and abnormalities of chromosomes 14 and 18. Trisomy 12 is considered an intermediate risk marker and a clonal driver mutation that occurs in early CLL evolution.Citation2,Citation21,Citation22 The detection of such abnormalities may change the choice of therapy and prognosis of the patients. The new cytogenetic abnormalities detected during the course of the disease in our study changed the prognostic classification of 86.4% of the patients conferring a worse prognosis in 58% and a better prognosis in 16% of them. Although karyotypic analysis is a better tool to investigate CE in CLL than FISH as it uncovers chromosomal aberrations through the whole genome, the application of both is also recommended as FISH can uncover more effectively submicroscopic abnormalities.
The parallel application of karyotype, metaphase and interphase FISH at diagnosis and during the course of CLL is recommended
Interphase FISH is a useful method for CLL, which is usually offered as a more sensitive test. In our study, FISH revealed a higher, although insignificantly, overall abnormality detection rate (69.05%) than CBA (62.4%). This higher rate was based on the ability of FISH to detect submicroscopic abnormalities, usually undetectable by karyotype. Deletions of 13q14.3 and 17p13, which are often cytogenetically cryptic, accounted almost entirely for the increased detection rate of iFISH. The ability of FISH to detect submicroscopic aberrations explains the fact that the most common aberration in FISH was del(13)(q14.3) compared to +12 in karyotype. For the same reason a higher frequency for del(17)(p13.1) and del(11)(q22.3) was also observed by FISH than karyotype.
However, FISH reveals chromosomal lesions restricted to specific chromosomal regions detected by the used probes and unfortunately its cost increases dramatically with the number of the used probes. Since karyotype after DSP-30/IL-2 stimulation can detect a variety of chromosomal abnormalities, providing additional information to FISH, which may alter the prognosis of FISH abnormalities as we showed in our patients, karyotypic analysis must always be performed. FISH should be performed in conjunction with karyotyping, to evaluate fast a large number of nuclei (∼200), to circumvent the lack of mitosis and to detect subtle abnormalities such as del(13q) that may be missed by CBA.
Diagnostic FISH analysis, almost always, is performed in interphase cells. In our study, we showed that mFISH gives more information than iFISH using the same CLL probes. This is due to its ability to visualize the location of a specific probe on the metaphase chromosomes. Consequently using mFISH we are able to see not only the loss or gain of specific chromosomal regions detected by the corresponding CLL probes but also if these chromosomal regions are translocated or inserted to other chromosomal regions. It is noteworthy that in our study 13q34 region was found translocated in three cases by mFISH. Translocations in CLL have been reported to confer poor prognosis although recent studies suggest that the poor outcome associated with translocations is in the context of a complex karyotype or an unbalanced translocation.Citation23–Citation25 Under this perspective mFISH has to be performed in parallel with iFISH in order to detect possible submicroscopic translocations or insertions which may have prognostic significance. Therefore mFISH is recommended in addition to iFISH on fixed cells after DSP-30/IL-2 stimulation so as to be able to study metaphases of CLL cells and not metaphases of normal cells which are much more common in unstimulated cultures.
Summary
In summary, our study demonstrates the selective advantage of CLL cells in DSP-30/IL-2 cultures and answers ambiguous issues of CLL. More specifically, we showed that DSP-30/IL-2 has an increased sensitivity for the detection of cytogenetically visible lesions compared to unstimulated cells, revealing more than 60 different chromosomal aberrations and provided additional information to FISH in 75.4% of patients. In most cases this information altered patients’ prognosis based on FISH results. We also proved that mFISH must be performed in addition to iFISH always on DSP-30/IL-2 stimulated cells so as to uncover submicroscopic translocations or insertions undetectable by iFISH. The comparison between DSP-30/IL-2 cultures and unstimulated cultures indicated that loss of chromosome Y is rather an age-related phenomenon and not a specific aberration of CLL. Moreover, we propose that karyotypic analysis should be performed not only at diagnosis but also during the course of the disease as CE detected in 50% of our cases with available follow-up karyotypic data, changed the prognosis influencing treatment choices of CLL patients. Therefore, the parallel application of karyotype, metaphase and interphase FISH after DSP-30/IL-2 stimulation is recommended for CLL investigation not only for diagnostic purposes but also for the investigation of CLL pathogenesis.
Disclaimer statements
Contributors None.
Funding None.
Conflicts of interest None.
Ethics approval Ethical approval was received for this study.
References
- Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–56. doi: 10.1182/blood-2007-06-093906
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al.. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. doi: 10.1056/NEJM200012283432602
- Muthusamy N, Breidenbach H, Andritsos L, Flynn J, Jones J, Ramanunni A, et al.. Enhanced detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide in combination with pokeweed mitogen and phorbol myristate acetate. Cancer Genet. 2011;204(2):77–83. doi: 10.1016/j.cancergen.2010.12.006
- Davids MS, Vartanov A, Werner L, Neuberg D, Dal Cin P, Brown JR. Controversial fluorescence in situ hybridization cytogenetic abnormalities in chronic lymphocytic leukaemia: new insights from a large cohort. Br J Haematol. 2015;170(5):694–703. doi: 10.1111/bjh.13498
- Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C. Immunostimulatory oligonucleotide-induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: a study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood. 2006;108(9):3152–60. doi: 10.1182/blood-2006-02-005322
- Liaw FP, Lau LC, Lim AS, Lim TH, Lee GY, Tien SL. CpG oligonucleotide and Interleukin 2 stimulation enables higher cytogenetic abnormality detection rates than 12-o-tetradecanolyphorbol-13-acetate in Asian patients with B-cell chronic lymphocytic leukemia (B-CLL). Int J Hematol. 2014;100(6):545–53. doi: 10.1007/s12185-014-1681-0
- Shanafelt TD, Hanson C, Dewald GW, Witzig TE, LaPlant B, Abrahamzon J, et al. Karyotype evolution on fluorescent in situ hybridization analysis is associated with short survival in patients with chronic lymphocytic leukemia and is related to CD49d expression. J Clin Oncol. 2008;26(14):e5–6. doi: 10.1200/JCO.2008.16.7874
- Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21(12):2442–51. doi: 10.1038/sj.leu.2404935
- Heerema NA, Byrd JC, Dal Cin PS, Dell’ Aquila ML, Koduru PR, Aviram A, et al. Stimulation of chronic lymphocytic leukemia cells with CpG oligodeoxynucleotide gives consistent karyotypic results among laboratories: a CLL Research Consortium (CRC) Study. Cancer Genet Cytogenet. 2010;203(2):134–40. doi: 10.1016/j.cancergencyto.2010.07.128
- Decker T, Schneller F, Sparwasser T, Tretter T, Lipford GB, Wagner H, et al. Immunostimulatory CpG-oligonucleotides cause proliferation, cytokine production, and an immunogenic phenotype in chronic lymphocytic leukemia B cells. Blood. 2000;95(3):999–1006.
- Takeshita F, Leifer CA, Gursel I, Ishii KJ, Takeshita S, Gursel M, et al. Cutting edge: role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J Immunol. 2001;167(7):3555–8. doi: 10.4049/jimmunol.167.7.3555
- Decker T, Bogner C, Oelsner M, Peschel C, Ringshausen I. Antiapoptotic effect of interleukin-2 (IL-2) in B-CLL cells with low and high affinity IL-2 receptors. Ann Hematol. 2010;89(11):1125–32. doi: 10.1007/s00277-010-0994-1
- Buhmann R, Kurzeder C, Rehklau J, Westhaus D, Bursch S, Hiddemann W, et al. CD40L stimulation enhances the ability of conventional metaphase cytogenetics to detect chromosome aberrations in B-cell chronic lymphocytic leukaemia cells. Br J Haematol. 2002;118(4):968–75. doi: 10.1046/j.1365-2141.2002.03719.x
- Atlas of Genetics and Cytogenetics in Oncology and Heamatology, atlasgeneticsoncology.org
- Chapiro E, Antony-Debre I, Marchay N, Parizot C, Lesty C, Cung HA, et al. Sex chromosome loss may represent a disease-associated clonal population in chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2013;53(3):240–7. doi: 10.1002/gcc.22134
- Karakosta M, Kalotychou V, Kostakis A, Pantelias G, Rombos I, Kouraklis G, et al. UGT1A1*28 polymorphism in chronic lymphocytic leukemia: the first investigation of the polymorphism in disease susceptibility and its specific cytogenetic abnormalities. Acta Haematol. 2013;132(1):59–67. doi: 10.1159/000355714
- Karakosta M, Tsakiridou A, Korantzis I, Manola KN. Deletion of 5q as a rare abnormality in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2010;200(2):175–9. doi: 10.1016/j.cancergencyto.2010.04.002
- Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. Cancer Genome Anatomy Project.
- Houldsworth J, Guttapalli A, Thodima V, Yan XJ, Mendiratta G, Zielonka T, et al. Genomic imbalance defines three prognostic groups for risk stratification of patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2014;55(4):920–8. doi: 10.3109/10428194.2013.845882
- Cavazzini F, Rizzotto L, Sofritti O, Daghia G, Cibien F, Martinelli S, et al. Clonal evolution including 14q32/IGH translocations in chronic lymphocytic leukemia: analysis of clinicobiologic correlations in 105 patients. Leuk Lymphoma. 2012;53(1):83–8. doi: 10.3109/10428194.2011.606384
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–26. doi: 10.1016/j.cell.2013.01.019
- Falisi E, Novella E, Visco C, Guercini N, Maura F, Giaretta I, et al. B-cell receptor configuration and mutational analysis of patients with chronic lymphocytic leukaemia and trisomy 12 reveal recurrent molecular abnormalities. Hematol Oncol. 2014;32(1):22–30. doi: 10.1002/hon.2086
- Mayr C, Speicher MR, Kofler DM, Buhmann R, Strehl J, Busch R, et al. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107(2):742–51. doi: 10.1182/blood-2005-05-2093
- Baliakas P, Iskas M, Gardiner A, Davis Z, Plevova K, Nguyen-Khac F, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89(3):249–55. doi: 10.1002/ajh.23618
- Van Den Neste E, Robin V, Francart J, Hagemeijer A, Stul M, Vandenberghe P, et al. Chromosomal translocations independently predict treatment failure, treatment-free survival and overall survival in B-cell chronic lymphocytic leukemia patients treated with cladribine. Leukemia. 2007;21(8):1715–22. doi: 10.1038/sj.leu.2404764
