Abstract
Current approaches based on electrophoretic, chromatographic or immunochemical principles have allowed characterizing multiple allergens, mapping their epitopes, studying their mechanisms of action, developing detection and diagnostic methods and therapeutic strategies for the food and pharmaceutical industry. However, some of the common structural features related to the allergenic potential of food proteins remain unknown, or the pathological mechanism of food allergy is not yet fully understood. In addition, it is also necessary to evaluate new allergens from novel protein sources that may pose a new risk for consumers. Technological development has allowed the expansion of advanced technologies for which their whole potential has not been entirely exploited and could provide novel contributions to still unexplored molecular traits underlying both the structure of food allergens and the mechanisms through which they sensitize or elicit adverse responses in human subjects, as well as improving analytical techniques for their detection. This review presents cutting-edge instrumental techniques recently applied when studying structural and functional aspects of proteins, mechanism of action and interaction between biomolecules. We also exemplify their role in the food allergy research and discuss their new possible applications in several areas of the food allergy field.
1. Introduction
Although any food can trigger anaphylaxis, fourteen of them (milk, egg, peanuts, tree nuts, fish, soybean, cereals containing gluten, crustaceans, mollusks, lupin, sesame, mustard, celery and sulphites), are mandatory on the labeling for safety reasons. However, aspects such as food processing and food matrix interactions should be considered to determine the allergenicity risk assessment. In addition, the use of new sources of dietary protein is increasing in recent years and its potential allergenicity needs to be evaluated to protect consumers (Verhoeckx et al. Citation2016). An exhaustive allergenicity risk assessment is needed before marketing novel food sources to evaluate the cross reactivity or de novo allergenicity of novel food proteins (Remington et al. Citation2018). However, the lack of cellular and animal models assays for detection of de novo sensitization are some of the identified gaps in the current allergenicity risk assessment that need to be further investigated.
Current approaches based on electrophoretic, chromatographic or immunochemical principles have allowed characterizing multiple allergens, mapping their epitopes, studying their mechanism of action, and developing detection and diagnostic methods for the food and pharmaceutical industry, as well as advancing therapeutic strategies. However, despite the high number of studies addressing the structure, digestibility and stability of food allergens against technological treatments, as well as the identification of the sequence of most of them (Aalberse Citation2000; Sirvent et al. Citation2009; Willison et al. Citation2011; Benedé et al. Citation2014; Villas-Boas et al. Citation2015; Benedé et al. Citation2015), no common biochemical properties have been associated with all allergenic food proteins, although few of them are shared (Costa et al. Citation2020; Costa et al. Citation2021).
Naturally occurring allergens are usually proteins, whose molecular weight needs to be at least 3.5 kDa to cross-link with two IgE molecules on the surface of mast cells and basophils (Bogh, Barkholt, and Madsen Citation2013). A molecular mass below 70 kDa is usually observed in food allergens (Vickery, Chin, and Burks Citation2011), although there is great variability in their sizes (Costa et al. Citation2020). Resistance to digestion and processing are regarded as common properties of food allergens. However, these characteristics do not provide sufficient information for allergenicity prediction because there are exceptions such as Ara h 2 from peanut (Koppelman et al. Citation2010) or Mal d 2 from apple (Smole et al. Citation2008). Processing has been used to modify allergenicity of proteins. For example, roasted peanuts are usually used for oral food challenge to confirm the suspicious of peanut allergy as well as for peanut oral immunotherapy in the current practice to reduce the adverse reactions caused by the used of raw peanuts. Oral immunotherapy using roasted peanut flour has been shown to be effective in desensitizing peanut-allergic children although a high number of adverse events have still been described (Anagnostou et al. Citation2014). Boiled peanuts are less allergenic than roasted peanuts (Beyer et al. Citation2001) and it has also been proposed to be used in oral immunotheraphy since boiling reduces IgE allergenicity while retaining T cell reactivity (Tao et al. Citation2016).
Susceptibility to proteolysis also depends on the food product and processing. In this respect, some epitopes may be destroyed and new epitopes may be formed (Thomas et al. Citation2007). In general, proteolysis potentially disrupts the protein structure, changing the structure of both the linear and the conformational epitopes present in food protein. However, the effect on allergenicity depends on the amino acid sequence and protein secondary structure (Thomas et al. Citation2007). In the case of peanuts, Ara h 2 hydrolysis has been demonstrated to change the secondary structure of the protein, without affecting its allergenicity due to the fact that linear epitopes are retained after proteolysis (Sathe, Teuber, and Roux Citation2005). In the case of milk, proteolysis reduces the number of epitopes that results in the reduction of allergenicity (Hochwallner et al. Citation2014). A high number of allergic children tolerated baked milk and egg due to the destruction of conformational epitopes, facilitating access to the digestive enzymes, and reducing intestinal absorption (Upton and Nowak-Wegrzyn Citation2018). However, the interactions between milk proteins and some components of the food matrix during heating seem to play the most important role in the reduction of allergenicity, limiting the accessibility of peptides to the immune system (Bavaro et al. Citation2019). In contrast, food matrix interactions may also increase the allergenicity of proteins, as it happens with roasting that increases allergenicity of Ara h 1 and Ara h 2 due to the neo-epitopes formation by Maillard Reaction (Maleki et al. Citation2000).
Stability toward proteolytic degradation and processing might differ between allergens depending also on structural properties. Amino acid composition of proteins determines physicochemical properties such as flexibility, solvent accessibility, solubility and electrostatic potential, which can influence their stability and their IgE binding capacity (Dall’ Antonia et al. Citation2014; Lozano-Ojalvo, Benedé, and van Bilsen Citation2021). For this reason, the evaluation of amino acid sequence homology of new proteins with already identified allergens is a currently applied approach to assess allergenicity and predict the allergenic potential of proteins, although it has been widely criticized (Silvanovich et al. Citation2006; Goodman et al. Citation2008; Cressman and Ladics Citation2009). In addition, many allergens undergo post-translational modifications that may influence protein fold and structure and, therefore, have an impact on protein allergenicity (Pekar, Ret, and Untersmayr Citation2018). Binding of ligands also leads to changes in allergen structure and stability against processing and proteolytic degradation (Pekar, Ret, and Untersmayr Citation2018) and they can act as immune-stimulators, thus inducing innate immune responses (Tordesillas et al. Citation2017). Therefore, the effect of the matrix in which allergens are usually consumed and the ability of the fragments generated upon gastrointestinal digestion to cross the intestinal barrier and interact with the gut-associated lymphoid tissue should also be evaluated (Lozano-Ojalvo, Benedé, and van Bilsen Citation2021). This will provide more data on how food processing and food matrix interaction modify the allergenicity of proteins. IgE-binding assays, applying sera from patients with established specific IgE response to the target food, are also used to evaluate the allergenicity of new proteins derived from a known allergenic food, but they are useless when the new protein is derived from a non-allergenic food (Mazzucchelli et al. Citation2018).
Given the current situation, more research is needed to find biochemical features that allow determining whether a protein has the potential to be allergenic. In this regard, the technological development has allowed the expansion of advanced technologies for which their full potential has not yet been used and could provide novel contributions to still unexplored molecular traits underlying both the structure of food allergens and the mechanism through which they sensitize or elicit an adverse response in human subjects, as well as improving analytical techniques for their detection.
In this review, we summarized the novel instrumental techniques often applied when studying structural and functional aspects of proteins, mechanisms of action and interaction between biomolecules. We further described these techniques and exemplify their current and perspective use in food allergy research. In addition, we discussed new possible applications of advanced analytical techniques in several areas of the food allergy field, including the structural characterization of allergens and mapping of epitopes, the study of posttranslational modifications, allergen detection and pathogenesis, diagnosis and treatment of the disease.
2. New techniques to determine structural and functional aspects of allergens
Three-dimensional structure determination of allergens is essential for analyzing exposed surface areas and mapping conformational epitopes. Moreover, it determines physicochemical properties such as flexibility, solvent accessibility, solubility and electrostatic potential, which can influence their IgE-binding capacity (Dall’ Antonia et al. Citation2014), stability against processing or enzymatic cleavage sites that are accessible to proteases (Abdullah et al. Citation2016). Protein tertiary structure may be investigated using several spectroscopic approaches. However, many molecular systems in cells that can interact with food allergens are either immobilized or motionally restricted, including membrane proteins, amyloid fibrils, and large molecular weight protein-protein or protein-nucleic acid complexes. Due to the lack of long-range order, these structures are resistant to crystallization and, therefore, create many difficulties in the application of high-resolution methods of X-ray crystallography, solution NMR or electron microscopy (Ladizhansky Citation2019). Therefore, new technological strategies have been developed to overcome this limitation.
2.1. X-ray free-electron lasers crystallography
X-Ray crystallography is the most powerful method to obtain information about the tertiary structure of proteins and protein complex at the atomic level. Briefly, the method is based on the diffraction of X-rays on protein crystals, data collection and model building.
Most allergens are relatively small, stable and well-structured proteins and, therefore, they are perfectly suited for X-ray crystallography studies. Numerous structures of food allergens have been solved and deposited in the Protein Data Bank. However, there are still difficulties in protein crystallography of large-sized molecules because this technique requires large well-ordered protein crystals that can diffract to high resolution with limited X-ray dose (Gallat et al. Citation2014). Big molecules rarely form such crystals, but rather generate nanometer-sized crystals, sensitive to radiation damage, with low-diffraction capabilities.
X-Ray free-electron lasers (FEL) opened new opportunities to overcome these drawbacks (Chapman et al. Citation2006). With an ultrashort and extremely bright coherent X-ray pulse, a single diffraction pattern may be recorded from a large macromolecule before the sample explodes and turns into a plasma. Sample damage by X-rays limits the resolution of structural studies on non-repetitive and non-reproducible structures such as individual biomolecules or cells. Cooling can slow sample deterioration, but cannot abolish damage-induced sample movement during the time needed for conventional measurements (Neutze et al. Citation2000). X-Ray FEL are expected to permit high-resolution diffractive images of nanometer- to micrometer-sized objects without the need for crystalline periodicity in the sample. Therefore, all large-size food allergens (i.g. homotrimeric Ara h 1) and the complexes of food allergens with IgE could be analyzed by X-ray FEL.
So far, this new method has not found a wide application in the characterization of food allergens. A singular example of the application of this technology for resolving the structure of a food allergen is the case of Gal d 4 (hen’s egg lysozyme). Its room-temperature crystal structures (Protein Data Bank ID: 3WUL) have been reported at a resolution better than 2 Å, using grease as a general carrier of protein microcrystals (Sugahara et al. Citation2015). The grease matrix-based approach should be applicable to a wide range of proteins, including single allergens and food allergen complexes in order to solve their structures at room temperature. Similar to X-ray crystallography, the method is applicable to purified proteins, and may be useful in risk assessment of novel foods, for which cross-reactivity of single protein to already known allergens is assessed based on homology and structural similarity at primary, secondary and tertiary levels (Mazzucchelli et al. Citation2018).
2.2. Solid-state nuclear magnetic resonance methods
Solid-state nuclear magnetic resonance (ssNMR) is (NMR) spectroscopic technique applied to condensed phase systems, including membrane proteins. This technique does not require crystals or fast molecular tumbling and, therefore, is not limited by molecular size. ssNMR can provide information at atomic level on insoluble systems, which are either disordered or lack long-range order (Sun et al. Citation2012; Wylie et al. Citation2016; Ladizhansky Citation2019). Structural and functional analyses are challenging, requiring new approaches to better characterize these systems. ssNMR is uniquely suited to the examination of membrane proteins in native environments and has the capability to elucidate complex protein mechanisms and structures (van der Wel Citation2017; Loquet et al. Citation2018; Ladizhansky Citation2019).
In the case of insoluble, high molecular weight, non-crystalline, and/or disordered food allergenic proteins (e.g. soybean hydrophobic seed protein), ssNMR can be used effectively for epitope characterization, including interactions between allergen and IgE, and especially hydrophobic natural ligands such as short-chain fatty acids, lipopolysaccharides, and phytosterols. ssNMR can provide information about the binding site and structure of protein-protein and protein-ligand interactions, accurate 3 D structure determination, and dynamical behavior of insoluble and/or disordered allergenic proteins (Dall’ Antonia et al. Citation2014). In addition, ssNMR can contribute to the prediction of cross-reactions between different food allergens as in the case of cross-reactivity between birch pollen and apple allergens, the interaction of allergens with solid matrices such as certain polysaccharides, and to the determination of the effect of food processing on the structure of allergen proteins (Krushelnitsky, Reichert, and Saalwachter Citation2013). However, although ssNMR is a structure-based prediction tool able to provide promising results on the detection of allergen structures, it has not yet been applied in this field. The closest application of ssNMR in the food allergen field regarded the determination of the purity of chitin obtained from ground shrimp shells to ensure the complete absence of allergens in the final product (King et al. Citation2017).
2.3. Cryo-electron microscopy coupled to mass spectrometry
Another emerging structural technique that could contribute to IgE epitope mapping is cryo-electron microscopy (cryo-EM). In this technique, the proteins do not need to be crystallized, but instead are instantly flash-frozen into vitrified ice, allowing to work with much larger biomolecular complexes and much less sample than the required for crystallography (Mueller Citation2017). Therefore, it opens an opportunity to directly examine the structure of an IgE molecule in complex with an allergen. In fact, cryo-EM has been used to map polyclonal epitopes of HIV immunized rabbits (Bianchi et al. Citation2018). This technique has also allowed studying the interior of the casein micelles and confirming the existence of inner cavities that are connected by wide channels (Trejo et al. Citation2011), which could explain its ability to retain macromolecules (Glab and Boratynski Citation2017). This singular structure allows casein micelles to act as carriers for other cow’s milk allergens, which could protect them against gastrointestinal digestion and technological treatments (Wang et al. Citation2017).
However, cryo-EM shows some limitations in providing high-resolution structures and, therefore, complementary techniques have been sought to solve this problem (Schmidt and Urlaub Citation2017). Stimulated by the latest developments in both instrumentation and image-processing software, chemical cross-linking combined with mass spectrometry (CX-MS) has evolved rapidly in recent years, consisting of introducing covalent bonds between two amino-acid residues in close proximity using a chemical crosslinking reagent. The cross-linked residues can be identified by mass spectrometry after proteolytic digestion (Leitner et al. Citation2016), allowing to identify regions that are non-covalently bound (van-der-Waals and electrostatic interactions) in interactions within and between proteins, which could be useful to provide advanced knowledge regarding the three-dimensional confirmation of food allergens.
Many food allergens show protein secondary structures, such as beta sheets, which impede cross-linking, as they are stabilized by hydrogen bonds and the reactive residues are no longer available for the crosslinking. Development of more unspecific and concomitantly cross-linkers could be an important direction of CX-MS-based structural research in the future (Schmidt and Urlaub Citation2017), expanding its application to the study of a large number of food allergenic proteins.
2.4. Bioinformatics
Structural similarity among proteins, as well as the correlation of their functions, are important to identify new allergenic proteins. In recent years, the development of bioinformatic tools to predict allergenicity has grown significantly and current methodologies use tools based not only on sequence identity, but also on the homology of functional motifs, which might retain the conformational epitope structure.
According to the guidelines from Food and Agriculture Organization of the United Nations and World Health Organization (FAO/WHO) to assess the potential allergenicity of genetically modified foods, cross-reactivity between a protein and a known allergen is established if they have a sequence similarity greater than 35% over a window of 80 amino acids, and if there are at least six contiguous exactly matching amino acids in both sequences. This report includes a variety of computational techniques mainly based on the comparison of protein sequences, although no indications on how these relationships were analyzed are given (Codex Alimentarius Commission Citation2001). Therefore, bioinformatics methods of allergenicity prediction can be grouped in three main groups: sequence, motif based approaches and support vector machine-based methods.
Sequence approaches are directly based on the requirements of WHO/FAO. They extract information from allergen databases and compare sequences using standard alignment tools (Stadler and Stadler Citation2003; Fiers et al. Citation2004; Goodman et al. Citation2016). However, the sequence identity between proteins does not always explain cross-reactivity. For example, Act d 11 from kiwifruit and Bet v 1 from birch pollen have a very low sequence identity, but both are recognized by the IgE of sera from birch pollen-allergic patients (Avino et al. Citation2011).
To increase the efficiency of these methods, the motif based-approaches appeared. They take into account, not only the sequence identity, but also the presence of characteristic motifs on the protein surface, such as B, T and IgE epitopes, the hydrophobic residues or structural similarity regions (Ivanciuc, Schein, and Braun Citation2003; Maurer-Stroh et al. Citation2019). These techniques help predicting cross-reactivity between allergens, though they have also been used for the identification of proteins with low allergenic potential in novel foods, which can be used for the development and validation of methods to assess proteins with allergenic potential (Krutz et al. Citation2019).
More sophisticated tools use Support Vector Machine-based (SVM) methods, which are statistical techniques that use input vectors from either amino acid composition or scores of pair-wise sequence similarity with known allergens, followed by the use of machine learning approaches to set up allergenicity (Cui et al. Citation2007; Muh, Tong, and Tammi Citation2009; Wang, Zhang, and Li Citation2013). Studies assessing the performance of these bioinformatic procedures show that motif based-approaches and SVM-based methods increase the accuracy and specificity of predictions (Wang et al. Citation2013; Maurer-Stroh et al. Citation2019). However, despite the soundness of current programs, the number of false positives in allergenicity predictions is still too high and other variables should be taken into account. For example, a significantly large number of food allergens bind and transport lipids whose ability to modulate the immune response has been reported (Tordesillas et al. Citation2017; López-Fandiño Citation2020). Therefore, it seems advisable to include other characteristics that could help assessing the real likelihood of proteins to be allergenic.
It should be taken into account that these methods are based on the similarity search of sequence or 3 D structure of proteins, so it is necessary the complete sequence. This feature is one of their main limitations since they are not suitable for identification of new allergenic proteins or proteins with unknown sequence. Despite this, it is important to remark that analyzing the proteome of novel foods can be done by means of proteomic analysis. This is the case of some recent advances related to important crops whose consumption has widely increased over the last few years, such as the characterization of soybean (Uniprot proteome ID UP000008827) or quinoa proteome (Capriotti et al. Citation2015). In this context, computational screening for potential allergenicity/IgE-cross-reactivity is both fast and inexpensive in contrast with in vitro experiments. Therefore, despite their limitations and the desire for further development, computational methods developed in the context of prediction of allergenicity are a useful tool that can be used in combination with other techniques in order to improve the risk assessment prediction.
3. Methods to decipher the mechanism of action of allergens in food mixtures without structural or functional characterization
Deciphering the signals that emerge from the cellular microenvironments could provide insights on the initiation of the cellular response to food allergens. Any initiation event starts with an interaction between the molecules of study and the biomolecules from the microenvironment. Therefore, improving our understanding of cellular responses to novel food allergens could arise from the application of recent methodologies developed to identify protein targets, predicting the mechanisms of action from an unbiased perspective. The food ingredient would resemble the complexity of chemical mixtures, that are complex bioactive samples that have been recently evaluated in toxicology to predict the impact of the exposure to complex mixtures of pollutants (Legler et al. Citation2020).
The current strategies to identify molecular targets have been focused on two directions: (1) phenotypic screening that provide direct observation of the cellular responses, but struggle with the identification of the molecular interactors (Moffat et al. Citation2017) and (2) screening that requires some structural information of the molecules, but may not be available as in the case of novel food allergens (Ziegler et al. Citation2013).
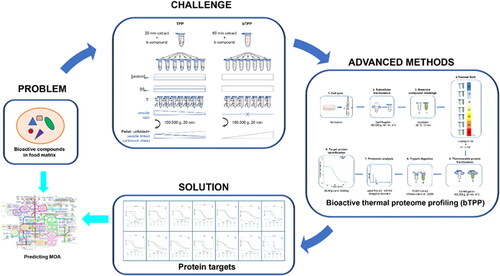
The first methodology that provided a more comprehensive identification of protein targets in a realistic cellular environment was the cellular thermal shift assay (CETSA) (Martinez Molina et al. Citation2013). This method measures the increase in thermal stability from cellular proteins due to their interaction with a bioactive compound and identifies the targets by immunoblotting, which is the limitation of the method (Vedadi et al. Citation2006; Martinez Molina et al. Citation2013).The next development was a method called thermal proteome profiling (TPP) that is also based on thermal shift assays, but the targets are identified by quantitative mass spectrometry (MS) based on isobaric tandem mass 10-plex reagents (Savitski et al. Citation2018). Soluble proteins from a cell lysate are incubated with or without the studied compound at a range of temperatures from 37 °C to 67 °C, being followed by centrifugation to separate soluble folded proteins from the thermally unfolded ones. The supernatants are analyzed by MS to generate the melting curves of the target proteins (Savitski et al. Citation2014; Franken et al. Citation2015). The method has been applied to study drug-target (Becher et al. Citation2016; Azimi et al. Citation2018), protein-substrate interaction in complex samples (Turkowsky et al. Citation2019) and protein degradation (Savitski et al. Citation2018). The main limitation of the TPP method was related with the fact that the analyzed soluble proteome contained a mixture of a soluble and vesicular fraction. Therefore, the TPP method could not guarantee a constant concentration of protein and compound at different temperatures, which is crucial to correctly calculate the melting point of the targets. This has recently been solved by the development of a new method called bioactive thermal proteome profiling (bTPP) that is based on a different postulate for protein solubility (Carrasco del Amor et al. Citation2019). The new method includes precipitation of the microsomal fraction before the thermal shift assay to ensure that the concentration of the available protein and compound is constant at any temperature (Carrasco del Amor et al. Citation2019) (). Moreover, this method does not require previous knowledge from the compound at the structural or functional level, and it is applicable to complex mixtures. Hence, it offers new opportunities to characterize novel food allergens that could interact in the cellular microenvironment, individually or integrated into complex and uncharacterized mixtures. Additional improvements have been achieved in this field with the method called proteome integral solubility alteration (PISA) that increases target specificity and high-throughput by measuring the integral under the melting curves of the protein targets (Gaetani et al. Citation2019). This approach eliminates the statistical uncertainties derived from fitting the thermal curves that is the base of the previous thermal proteome profiling methodologies.
Figure 1. Graphical representation of bioactive thermal proteome profiling strategy. Problem: uncharacterized bioactive compound in food matrix; Challenge: to obtain a proteome-wide food bioactive compound interaction map; Advanced method: bioactive thermal proteome profiling; Solution: unbiased identification of protein targets by MS; Opportunity: prediction of mechanisms of action of a novel compound by one single experiment and analytical technique.
4. Techniques to identify post-translational modifications of allergens
Biologically occurring post-translational modifications (PTM) are chemical alterations (induced by enzymes) that proteins may undergo after biosynthesis, being critically relevant in regulating their structure and function (Lanucara and Eyers Citation2013). Additionally, PTM often determine the interactions among proteins and establish the basis for several cellular signaling pathways, playing crucial roles in immune modulation, inflammation, host-pathogen interactions, and degenerative/proliferative disorders (Pascovici et al. Citation2018). The most common PTM include the specific cleavage of precursor proteins, the formation of disulfide bonds, the covalent addition or removal of low-molecular-weight groups, which occur in the amino acid side chains and are generally catalyzed by enzymes (Khoury, Baliban, and Floudas Citation2011; Ravikiran and Mahalakshmi Citation2014). Particularly, allergens can suffer single or multiple biological PTM, such as glycosylation, phosphorylation, acetylation, hydroxylation, and methylation (Pekar, Ret, and Untersmayr Citation2018; Costa et al. Citation2020, Citation2021). Process-induced PTM of allergens can also occur by chemical modifications during food processing, namely deamidation or glycation (Maillard reactions) (Le, Deeth, and Larsen Citation2017). Despite being often used indiscriminately, biological- or process-induced PTM are two complete different and concepts because the first relates to PTM occurring during protein synthesis and the second results from food processing.
Currently, mass spectrometry techniques (by means of bottom-up, top-down and middle-down strategies) are able to evaluate the status of protein modification, including their site location (Olsen and Mann Citation2013; Pascovici et al. Citation2018). The bottom-up MS approach consists of analyzing the resulting peptides (0.8–3 kDa) from previous enzymatic or chemical cleavage and it has been widely used for qualitative analysis of new PTM (Lanucara and Eyers Citation2013; Pandeswari and Sabareesh Citation2019). This bottom-up MS approach is a very suitable technique to identify, locate and quantify modified peptides, although it has some limitations mainly concerning the combinatorial effect of several PTM. Working at a protein level, the top-down approach has been used to provide the number, position and type of PTM on a single polypeptide chain, offering an ideal analytical strategy for discovering the combinatorial effects of PTM. However, the requirements for its use are particularly challenging due to the size of the analytes (>10 kDa). The new middle-down approach has emerged as an alternative, also involving proteolysis, but in a restricted manner, producing longer proteolytic peptides (2.5–10 kDa) and a better sequence coverage (Pandeswari and Sabareesh Citation2019).
Most of the MS-based methods established for PTM analysis often target a single modification type, such as phosphorylation or glycosylation, and one type of sample/tissue (Pascovici et al. Citation2018). However, there are numerous proteins/allergens including several PTM sites and/or two or more different PTM (e.g. milk caseins). Significant advances have been carried out in PTM analysis, establishing the basis for developing proficient PTM workflows. Those include the creation of efficient sample preparation protocols (accurate selection of lysis and digestion protocols), as well as strategies for PTM enhancement (effective purification methods based on affinity or immunoprecipitation enrichment), and highly sensitive MS platforms for PTM quantification. Nonetheless, such PTM workflows are not easy to establish because they normally require the use of large amounts of starting materials (mg of proteins/peptides) that, depending on the sample/tissue, might be difficult to obtain (Pascovici et al. Citation2018). Additionally, the analysis of some PTM (e.g. phosphorylation, glycosylation) is a hard and challenging task due to a number of reasons. For instance, the low amounts of phosphorylated proteins, the challenges in isolating selective phosphopeptides and the peculiar chemistry of each phosphorylated amino acid are frequently accountable for the difficulties in analyzing phosphorylation as a PTM (Yakubu, Nieves, and Weiss Citation2019). Besides, glycosylation and phosphorylation modification sites are often difficult to identify since they are labile and can be lost during the separation and fragmentation process (Couto et al. Citation2018).
Glycosylation is one of the most relevant PTM in allergens, which can be identified by bottom-up and top-down approaches. Using the bottom-up approach, the application of MALDI-TOF/MS for the identification of potential glycosylation sites of Cor a 11 (hazelnut) has been described (Lauer et al. Citation2004). Similarly, the N-glycoforms of soybean allergenic glycoproteins were identified and quantified by ESI-MS, MS/MS and LC-MS/MS (Li et al. Citation2016). Phosphorylation, hydroxylation, acetylation and methylation are other important PTM associated to the allergenicity of proteins, which have been characterized by MS using the bottom-up approach. Barral et al. (Citation2005) described the existence of a phosphorylated serine residue at the N-terminal of Ole e 10 (olive) by MALDI-TOF analysis. Li et al. (Citation2009) characterized four isoforms of Ara h 2 (peanut) by LC-MS/MS and identified site‐specific hydroxylation of proline residues in each isoform. Lipid transfer proteins (LTP) were purified from the pulp and peel of different peach genotypes and analyzed by MALDI-TOF-MS/MS, revealing the presence of two methylated sites on the arginine residues in all LTP from peel (Larocca et al. Citation2013). Using a bottom‐up MS approach, Rahman et al. (Citation2010) analyzed the tropomyosin isolated from snow crab (Chionoecetes opilio) and demonstrated the existence of a N‐terminal acetylation as the site for the PTM (Rahman et al. Citation2010). Glycosylation and phosphorylation sites in κ-caseins of cow’s milk were characterized by two-dimensional gel electrophoresis coupled with MALDI-TOF-MS and nano ESI-MS/MS (Holland, Deeth, and Alewood Citation2004, Citation2006). Applying middle-down and top-down approaches on the subunit and at the protein level of Sin a 1 (mustard), Hummel, Wigger, and Brockmeyer (Citation2015) detected eight consensus isoforms. Despite not identifying specific PTM, different modifications in the isoforms and phytic acid as a potential ligand for Sin a 1 were reported. Bromilow et al. (Citation2017) exploited a combination of platforms to investigate the proteomic profiling of wheat gluten, using a QTOF instrument with a data-independent schema, which incorporated ion mobility (DIA-IM-MS) and a data-dependent acquisition (DDA) workflow, employing a linear ion trap quadrupole (LTQ) instrument. As example of process-induced PTM, the authors observed a widespread glutamine deamidation in all the different types of characterized gluten proteins.
Recent advances in MS-based approaches have shade some light regarding the composition and location of different PTM in several known allergens. Proteins with specific PTM have been related with increased allergenic potential, although the real impact of PTM in protein allergenicity is still unknown (Costa et al. Citation2020, Citation2021). However, it is also important to highlight that the application of MS-approaches in detecting PTM in novel food proteins might help establishing some relation to their allergenic potential.
Most of the studies (if not all) are performed on purified proteins, which restrains extrapolating the concrete impact of modified allergenic proteins within a food matrix. In terms of allergenicity risk assessment, the information retrieved from studies evaluating the PTM on specific proteins is still very limited, but integrating this knowledge with other allergen physicochemical properties might contribute to a better allergenicity risk assessment.
5. Methods to detect interactions between allergens and biomolecules
5.1. Mass spectrometry
According to the general mechanisms of IgE-mediated allergies, antigenic epitopes bind non-covalently IgE, forming IgE-antigen complexes that fit into a high-affinity receptor (FcεRIα) on the surface of mast cells and basophils. Cross-linking of FcεRIα-bound IgEs through multivalent allergens triggers the allergic cascade. Some structural aspects of IgE-FcεRIα interaction also remain controversial (Hirano et al. Citation2018). The fragmentary knowledge about the actual antigenic determinants and their structural interaction between with antibody isotypes or with receptors of immunocompetent cells is among the main factors preventing us to establish what makes a protein a food allergen. The incomplete picture of such a complex interplay hinders the process of allergenicity risk assessment, especially for novel potential allergens, as well as the possibility of devising novel therapeutic strategies.
Mass spectrometry (MS) is the core technology of several cutting-edge strategies that have been successfully devised to map protein domains involved in non-covalent interactions and expectedly will replace or, at least, support the existing methods in the near future.
Hydrogen/deuterium exchange (HDX) is an emerging MS-based epitope mapping method, which exploits the labeling of protein amide backbone with deuterium at variable rates, depending on the accessibility to solvent and on the local topology of the amino acid sequences. Antigens alone or antigen-antibody complexes are incubated in D2O, followed by proteolysis and analysis by LC-MS/MS. The domains involved in the antibody binding are those differing in the deuterium uptake and can be identified due to a molecular weight shift. Recently, the conformational epitopes of Ana o 2 (Zhang et al. Citation2013), Pru du 6 (Willison et al. Citation2013) and Ana o 1 (Guan et al. Citation2015) recognized by monoclonal antibodies have been mapped using this technique. This information could be useful to predict the epitope structure against human IgE and, with some adjustments, this technique can be in principle extended to the study of epitopic regions involved in the binding to specific IgE populations (Matsuo, Yokooji, and Taogoshi Citation2015). Related to the HDX-MS method, the chemical footprint by oxidative labeling of amino acid residues not involved in the interaction, namely Fast Photochemical Oxidation of Proteins (FPOP)-MS, is an emerging strategy to elucidate the structural details of protein-protein and protein-ligand interactions, also including the contacting epitope-paratope domains (Yan et al. Citation2014).
Polypeptide chains in close spatial proximity can be chemically cross-linked using a variety of commercially available bifunctional reagents, hooking specific amino acid residues (Leitner Citation2016). Upon proteolysis, cross-linked heterodimers are released from the former interacting protein domains and are identified by MS/MS. However, due to the difficult identification of peptides involved in the cross-link, this approach has not yet entered the routine practice for the epitope recognition.
The recently optimized limited proteolysis-coupled to MS (LiP-MS), which is a proteome-wide scale advance of a strategy developed more than 30 years ago (Jemmerson and Paterson Citation1986), addresses the functional analysis of protein interactions either with ligands or physiological biomolecular effectors (Schopper et al. Citation2017). The binding of a protein to a ligand or the interaction between proteins results in altered structural conformations of the polypeptide, producing different sets of peptides upon limited proteolysis, which are further hydrolyzed by trypsin. Combining the LC-MS analysis to a software-assisted reconstruction, it is possible to identify the protein regions involved in binding events. Recently, a workflow for quantitative chemoproteomic analysis has been developed (Piazza et al. Citation2018). This method enables the identification of metabolite-protein interactions and the functional modulation of allergen immunogenicity/allergenicity directly in their native environment. In addition, MS also enables the profiling of the entire repertoire of major histocompatibility complex-binding peptides exposed on the surface of the antigen-presenting cells (Bozzacco et al. Citation2011; Clement et al. Citation2016).
It is envisaged that the routine applications of these strategies to food allergen characterization will provide insights into the etiopathological mechanisms of the disease at a molecular level and will prime the development of adequate therapeutic approaches.
5.2. Label-free interaction analysis
Label-free detection methods utilize molecular biophysical properties such as the refractive index to monitor molecular interactions that are transduced as mechanical, electrical, or optical signals, acquiring direct information from native proteins and ligands (Ray, Mehta, and Srivastava Citation2010). They would replace current assays based on fluorescence, luminescence or radioactivity, which use specialized reagents labeled with detection moieties that can introduce artifacts. Any foreign molecule that is chemically or temporarily attached to the molecule of interest can potentially alter its intrinsic properties, changing its interaction capacity and, thus, generating false positives and negative results. Moreover, label detection techniques require a previous labeling process combining synthesis and purification that is usually low yield (Syahir et al. Citation2015). Label-free detection methods such as Surface Plasmon Resonance (SPR), microfluidics and biosensors are quantitative techniques that show in general higher sensitivity, lower cost, and greater ease of manufacture than traditional methods and can be applied for the analysis of allergens as contaminants in order to facilitate the correct food allergen labeling.
5.2.1. Surface plasmon resonance
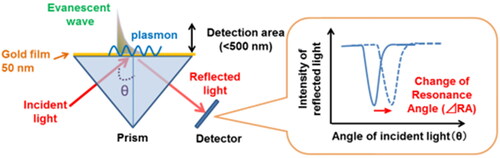
SPR refers to the electromagnetic response obtained when collective oscillations of free electrons (plasmons) occur on the surface of a metal. SPR takes place when polarized light strikes an electrically conducting surface, generally a gold layer, at the interface between two media with different refractive index, the sensor surface containing the immobilized proteins and a sample containing a potential interacting partner in buffer solution material (Sipova and Homola Citation2013) (). During the interaction, the angle of minimum intensity reflected light is detected, changing as molecules bind and dissociate. This technique allows studying the interactions between biomolecules in real-time and can be performed on complex mixtures, such as cell culture supernatants or cell extracts, working equally well on clear and colored or opaque samples (Michel, Xiao, and Alameh Citation2017).
Figure 2. Principle of surface plasmon resonance sensor. Surface plasmon resonance sensors detect a refractive index change within a detection area (<500 nm) as a modification of resonance angle (RA) (Yanase et al. Citation2014).
In the allergy field, SPR can be used to detect and measure the avidity of antibodies recognizing allergens (Chardin et al. Citation2014). The study of the influence of immunotherapy against the major birch pollen allergen, Bet v 1, on specific IgE, IgG1 and IgG4 affinities was one of the first applications of this technique that allowed to measure the IgG/IgE ratio in patients undergoing immunotherapy (Jakobsen et al. Citation2005).
It can also be useful to characterize polyclonal (Pol et al. Citation2007) and monoclonal (Laffer et al. Citation2008) anti-IgE to be used in immunotherapy against allergy and asthma. Antibody responses, induced in monkeys by recombinant IgE‐derived immunotherapeutic protein against atopic allergies and asthma, were characterized by SPR (Pol et al. Citation2007). This technique was also used to assess the binding kinetics and affinity of a monoclonal anti-IgE and investigate its potential use for depletion of IgE and isolation of IgE‐bearing cells from peripheral blood (Laffer et al. Citation2008). Another application of SPR has been the improvement of allergy diagnosis, reducing the detection limit in the determination of serum IgE in allergic patients (Hamilton, Saini, and MacGlashan Citation2012; Joshi et al. Citation2014) or visualizing the effect of various stimuli or inhibitors on basophils in a high throughput screening system (Yanase et al. Citation2012).
Due to the high sensitivity, low cost, and easy fabrication, SPR provide real-time methods to monitor the presence of allergenic ingredients in food matrices during processing (Zhou et al. Citation2019), although it should be noted that the ligand may not maintain its native configuration upon immobilization on the sensor chip surface. Simple and label-free SPR sensors have been develop to detect casein (Hiep et al. Citation2007), β-lactoglobulin (Ashley et al. Citation2018; Wu et al. Citation2016), ovalbumin (Pilolli, Visconti, and Monaci Citation2015; Lin et al. Citation2017), lysozyme (Ocana et al. Citation2015), Ara h 1 (Wu et al. Citation2016), parvalbumin (Lu, Ohshima, and Ushio Citation2004) and tropomyosin (Zhou et al. Citation2020). In addition, SPR biosensors allow the capture of multi-allergens in a single food sample for high-throughput multiplex analysis, as it was demonstrated by Billakanti et al. (Citation2010), who detected five whey proteins in both raw and processed milk.
5.2.2. Microfluidics
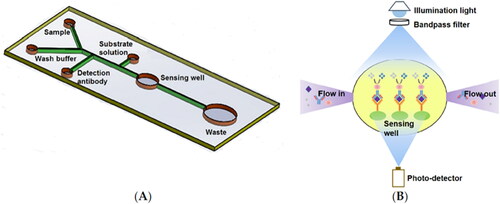
Microfluidics, the science of fluids on the micro- and nanometer scale, has become an increasingly popular tool in biochemistry applications (Cho et al. Citation2011). The integration of microfluidic systems in other technologies, such as biosensors or microarrays, can be useful in the field of food allergy. Microfluidic ELISA-based optical sensor has been applied for rapid detection of Ara h 1, reducing the total assay time from hours to 15–20 min and decreasing the sample/reagent consumption compared to commercial ELISA kits, with superior sensitivity (Weng, Gaur, and Neethirajan Citation2016) (). Besides, a microarray coupled to microfluidics has also been reported for the in vitro serological detection of specific IgE for allergy diagnosis (Cretich et al. Citation2009).
Figure 3. Microfluidic sensor. Schematic representation of the microfluidic ELISA sensor layout (not to scale) (A) and principle of the absorption detection with a miniaturized optical sensor device (B) (Weng, Gaur, and Neethirajan Citation2016).
5.2.3. Biosensors
The development of electrochemical biosensors for applications in food allergen analysis has increased in recent years. They are powerful tools to perform quick analyses with high sensitivity and selectivity, while allowing on-site analysis, although they generally require expensive instruments and skilled operators. They can be categorized as potentiometric, amperometric, voltammetric, conductometric or impedimetric, sensors according to the electrochemical principle involved. The use of specific antibodies has allowed the development of immunosensors for the selective detection of food allergenic proteins from peanut (Singh et al. Citation2010; Alves et al. Citation2015; V. R. V.Montiel et al. Citation2016), milk (Montiel et al. Citation2015; Inaba, Kuramitz, and Sugawara Citation2016; R. V. R. Montiel et al. Citation2016), egg (Čadková et al. Citation2015; Benedé et al. Citation2018), shrimp (Angulo-Ibañez et al. Citation2019), tomato (Pereira-Barros et al. Citation2019) or hazelnut (Montiel et al. Citation2017). However, limitations due to the use of antibodies such as nonspecific binding or the liability to degrade should be considered in immunosensor development.
6. Techniques to unravel the phenotype of single cells and novel immune pathways in food allergy
Beyond the technological advances made in the comprehensive investigation of allergen structure and characterization, the development of high-throughput “-omics” techniques (proteomics, transcriptomics, metabolomics, genomics, and epigenomics) has also allowed the progress of sophisticated approaches for studying biological pathways involved in food allergy through characterization of single cells (Dhondalay et al. Citation2018).
High-throughput proteomics has been used to study the allergenic response at single-cell resolution by applying mass cytometry or cytometry by time-of-flight (CyTOF). In contrast to fluorescent-based flow cytometry, that uses dye-labeled antibodies, CyTOF is a MS-based flow cytometry approach that uses isotopically purified heavy metal atoms as reporters (rather than fluorochromes), which are identified by a MS-TOF allowing the detection of molecules expressed by immune cells. Conventional fluorescent dye-based flow cytometry distinguishes up to 18 proteins at a single-cell level. However, CyTOF accurately differentiates more than 40 targets (theoretically up to 100 isotopes) with less cross-talk between detector channels and lesser signal overlap (Spitzer and Nolan Citation2016; Simoni et al. Citation2018). Based on this technology, imaging mass cytometry (IMC) has been recently applied to analyze tissue sections, being reviewed by Chang et al. (Citation2017). Some studies have revealed the importance of CyTOF for research and clinical monitoring of patients with food allergies. High-dimensional analyses of immune biomarkers using mass cytometry have been employed to study the diversity of T-cell phenotypes in food allergic patients (Chinthrajah et al. Citation2018) and to identify discrete B-cell subsets in subjects with red meat allergy (Cox et al. Citation2019). In addition, Tordesillas et al. (Citation2016) used CyTOF to describe a potential interaction between basophils and platelets after peanut exposure. Moreover, this technique has proven to be a robust approach to basophil activation testing that could improve the diagnosis and prognosis of food allergies (Mukai et al. Citation2017). CyTOF has also been applied for the investigation of a non-IgE mediated allergy, the food protein-induced enterocolitis syndrome (FPIES), and the results obtained have revealed a systemic immune activation following elicitation by foods (Goswami et al. Citation2017). In general, the current challenge in this field is the development of dimensionality reduction methods that allow examining, visualizing and presenting the vast amount of data generated by CyTOF.
Besides proteomics, transcriptomics is now being applied to better understand the complex mechanisms involved in food allergic responses by measuring transcriptionally active genes expressed by the immune cells (Dhondalay et al. Citation2018). Microarrays (platforms with probes to measure a set of predefined genes) are commonly used for the characterization of RNA molecules. However, the next-generation platform for gene expression analysis is RNA sequencing (RNA-seq). This approach is capable of analyzing the increasing flux of gene expression along with high-throughput bioinformatics. Compared to microarrays, RNA-seq has several advantages, including a higher percentage of differentially expressed genes and its ability to detect novel transcripts.
So far, it is considered that the genes expressed by immune cells are linked to allergic responses triggered by foods, although transcript levels are not always well correlated with released proteins (Dhondalay et al. Citation2018). Kosoy et al. (Citation2016) reported the transcriptional profiling of peripheral blood mononuclear cells (PBMC) from reactive individuals to egg proteins, showing an increase expression of genes associated with allergic inflammation and higher secretion of specific immune mediators compare to those from PBMCs from healthy donors. Using RNA-seq, Watson et al. identified variations in gene expression during acute allergic responses triggered by peanut (Watson et al. Citation2017). Moreover, this study allowed the recognition of key driver genes, biological processes and cell types associated with severe peanut allergic reactions. Recently, Croote et al. (Citation2018) performed plate-based single-cell RNA-seq on IgE-producing B-cells isolated from PBMC of food-allergic subjects to clarify the gene expression and splicing patterns of these cells. Accordingly, these antibodies attained high affinity with unpredicted cross-reactivity to peanut allergens (Ara h 2/Ara h 3), properties resulting from mutations in the variable regions of heavy and light chains of IgE in allergic individuals (Croote et al. Citation2018). Additionally, transcriptomics has also been employed to better comprehend food allergens and the genes involved in their expression/regulation (Baar et al. Citation2014; Ramesh et al. Citation2016; Nugraha et al. Citation2018). Therefore, transcriptomics data have already highlighted variances in the gene expression patterns of allergic and non-allergic subjects, revealing a strong potential to detect novel immune processes in food allergy (Dhondalay et al. Citation2018).
Finally, a new technology that combines proteomics and transcriptomics has emerged. The cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) connects the measurement of specific protein markers with the unbiased genome-wide expression profiling. This approach uses oligonucleotide-labeled antibodies to integrate cellular proteins and transcriptome measurements into an efficient, single-cell readout (Stoeckius et al. Citation2017). CITE-seq has not been used in studies of the allergic responses yet, but its application may unravel pathways involved in the pathogenesis of food allergy.
7. Conclusions
Technological development has allowed the expansion of advanced technologies that could be applied in food allergy research to decipher the structure of food allergens and the mechanisms through which they sensitize or elicit adverse responses in some individuals, as well as improving current analytical techniques for allergen detection (). Promising techniques such as X-ray FEL crystallography, ssNMR, cryo-EM/CX-MS or computational predictive methods could determine structural and functional aspects of allergens. In order to decipher the mechanisms of action of allergens without structural or functional characterization, the TPP approach could be useful. MS-based novel approaches could help identifying post-translational modifications of allergens and together with label-free interaction analysis to detect interactions between allergens and other biomolecules. In addition, techniques such as CyTOF, IMC, RNA-seq and CITE-seq can contribute to unravel the phenotype of single cells and novel immune pathways in food allergy.
Table 1. Novel advanced methods for application in the food allergy field.
Due to the fast technological advances, new insights into food allergy research need to be considered. The different approaches collected in this review probably offer the best or promising options to move forward in the food allergy field, to understand what turns a protein into an allergen, what underlies the allergic reaction, and how it can be abolished, in the not too distant future.
Disclosure statement
The authors declare that they have no conflicts of interests.
Additional information
Funding
References
- Aalberse, R. C. 2000. Structural biology of allergens. Journal of Allergy and Clinical Immunology 106 (2):228–38. doi: 10.1067/mai.2000.108434.
- Abdullah, S. U., Y. Alexeev, P. E. Johnson, N. M. Rigby, A. R. Mackie, B. Dhaliwal, and E. N. C. Mills. 2016. Ligand binding to an allergenic lipid transfer protein enhances conformational flexibility resulting in an increase in susceptibility to gastroduodenal proteolysis. Scientific Reports 6 (1):30279. doi: 10.1038/srep30279.
- Alves, R. C., F. B. Pimentel, H. P. A. Nouws, W. Correr, M. B. Gonzalez-Garcia, M. Oliveira, and C. Delerue-Matos. 2015. Detection of the peanut allergen Ara h 6 in foodstuffs using a voltammetric biosensing approach. Analytical and Bioanalytical Chemistry 407 (23):7157–63. doi: 10.1007/s00216-015-8879-8.
- Anagnostou, K., S. Islam, Y. King, L. Foley, L. Pasea, S. Bond, C. Palmer, J. Deighton, P. Ewan, and A. Clark. 2014. Assessing the efficacy of oral immunotherapy for the desensitisation of peanut allergy in children (STOP II): A phase 2 randomised controlled trial. The Lancet 383 (9925):1297–304. doi: 10.1016/S0140-6736(13)62301-6.
- Angulo-Ibañez, A., U. Eletxigerra, X. Lasheras, S. Campuzano, and S. Merino. 2019. Electrochemical tropomyosin allergen immunosensor for complex food matrix analysis. Analytica Chimica Acta 1079:94–102. doi: 10.1016/j.aca.2019.06.030.
- Ashley, J., R. D’Aurelio, M. Piekarska, J. Temblay, M. Pleasants, L. Trinh, T. L. Rodgers, and I. E. Tothill. 2018. Development of a beta-Lactoglobulin sensor based on SPR for milk allergens detection. Biosensors 8 (2):32. doi: 10.3390/bios80200.
- Avino, R., M. L. Bernardi, M. Wallner, P. Palazzo, L. Camardella, L. Tuppo, C. Alessandri, H. Breiteneder, F. Ferreira, M. A. Ciardiello, et al. 2011. Kiwifruit Act d 11 is the first member of the ripening-related protein family identified as an allergen. Allergy 66 (7):870–7. doi: 10.1111/j.1398-9995.2011.02555.x.
- Azimi, A., S. Caramuta, B. Seashore-Ludlow, J. Boström, J. L. Robinson, F. Edfors, R. Tuominen, K. Kemper, O. Krijgsman, D. S. Peeper, et al. 2018. Targeting CDK2 overcomes melanoma resistance against BRAF and Hsp90 inhibitors. Molecular Systems Biology 14 (3):e7858. doi: 10.15252/msb.20177858.
- Baar, A., S. Pahr, C. Constantin, S. Giavi, N. G. Papadopoulos, A. S. Pelkonen, M. J. Mäkelä, S. Scheiblhofer, J. Thalhamer, M. Weber, et al. 2014. The high molecular weight glutenin subunit Bx7 allergen from wheat contains repetitive IgE epitopes. Allergy 69 (10):1316–23. doi: 10.1111/all.12464.
- Barral, P., E. Batanero, M. Villalba, and R. Rodríguez. 2005. Expression of the major olive pollen allergen Ole e 10 in the yeast Pichia pastoris: Evidence of post-translational modifications. Protein Expression and Purification 44 (2):147–54. doi: 10.1016/j.pep.2005.04.012.
- Bavaro, S. L., E. De Angelis, S. Barni, R. Pilolli, F. Mori, E. M. Novembre, and L. Monaci. 2019. Modulation of milk allergenicity by baking milk in foods: A proteomic investigation. Nutrients 11 (7):1536. doi: 10.3390/nu11071536.
- Becher, I., T. Werner, C. Doce, E. A. Zaal, I. Tögel, C. A. Khan, A. Rueger, M. Muelbaier, E. Salzer, C. R. Berkers, et al. 2016. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nature Chemical Biology 12 (11):908–10. doi: 10.1038/nchembio.2185.
- Benedé, S., I. López-Expósito, R. López-Fandiño, and E. Molina. 2014. Identification of IgE-binding peptides in hen egg ovalbumin digested in vitro with human and simulated gastroduodenal fluids. Journal of Agricultural and Food Chemistry 62 (1):152–8. doi: 10.1021/jf404226w.
- Benedé, S., I. López-Expósito, E. Molina, and R. López-Fandiño. 2015. Egg proteins as allergens and the effects of the food matrix and processing. Food & Function 6 (3):694–713. doi: 10.1039/C4FO01104J.
- Benedé, S., V. R. V. Montiel, E. Povedano, M. Villalba, L. Mata, P. Galan-Malo, R. M. Torrente-Rodriguez, E. Vargas, A. J. Reviejo, S. Campuzano, et al. 2018. Fast amperometric immunoplatform for ovomucoid traces determination in fresh and baked foods. Sensors and Actuators B: Chemical 265:421–8. doi: 10.1016/j.snb.2018.03.075.
- Beyer, K., E. Morrow, X. M. Li, L. Bardina, G. A. Bannon, A. W. Burks, and H. A. Sampson. 2001. Effects of cooking methods on peanut allergenicity. Journal of Allergy and Clinical Immunology 107 (6):1077–81. doi: 10.1067/mai.2001.115480.
- Bianchi, M., H. L. Turner, B. Nogal, C. A. Cottrell, D. Oyen, M. Pauthner, R. Bastidas, R. Nedellec, L. E. McCoy, I. A. Wilson, et al. 2018. Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity 49 (2):288–300. doi: 10.1016/j.immuni.2018.07.009.
- Billakanti, J. M., C. J. Fee, F. R. Lane, A. S. Kash, and R. Fredericks. 2010. Simultaneous, quantitative detection of five whey proteins in multiple samples by surface plasmon resonance. International Dairy Journal 20 (2):96–105. doi: 10.1016/j.idairyj.2009.08.008.
- Bogh, K. L., V. Barkholt, and C. B. Madsen. 2013. The sensitising capacity of intact beta-lactoglobulin is reduced by co-administration with digested beta-lactoglobulin. International Archives of Allergy and Immunology 161 (1):21–36. doi: 10.1159/000343042.
- Bozzacco, L., H. Yu, H. A. Zebroski, J. Dengjel, H. Deng, S. Mojsov, and R. M. Steinman. 2011. Mass spectrometry analysis and quantitation of peptides presented on the MHC II molecules of mouse spleen dendritic cells. Journal of Proteome Research 10 (11):5016–30. doi: 10.1021/pr200503g.
- Bromilow, S. N. L., L. A. Gethings, J. I. Langridge, P. R. Shewry, M. Buckley, M. J. Bromley, and E. N. C. Mills. 2017. Comprehensive proteomic profiling of wheat gluten using a combination of data-independent and data-dependent acquisition. Frontiers in Plant Science 7. doi: 10.3389/fpls.2016.02020.
- Čadková, M., R. Metelka, L. Holubová, D. Horák, V. Dvořáková, Z. Bílková, and L. Korecká. 2015. Magnetic beads-based electrochemical immunosensor for monitoring allergenic food proteins. Analytical Biochemistry 484:4–8. doi: 10.1016/j.ab.2015.04.037.
- Capriotti, A. L., C. Cavaliere, S. Piovesana, S. Stampachiacchiere, S. Ventura, R. Zenezini Chiozzi, and A. Laganà. 2015. Characterization of quinoa seed proteome combining different protein precipitation techniques: Improvement of knowledge of non-model plant proteomics. Journal of Separation Science 38 (6):1017–25. doi: 10.1002/jssc.201401319.
- Carrasco del Amor, A., S. Freitas, R. Urbatzka, O. Fresnedo, and S. Cristobal. 2019. Deciphering mechanism of actions of novel compounds by bioactive thermal proteome profiling. Marine Drugs 17 (6):371. doi: 10.3390/md17060371.
- Chang, Q., O. I. Ornatsky, I. Siddiqui, A. Loboda, V. I. Baranov, and D. W. Hedley. 2017. Imaging mass cytometry. Cytometry Part A 91 (2):160–9. doi: 10.1002/cyto.a.23053.
- Chapman, H. N., A. Barty, M. J. Bogan, S. Boutet, M. Frank, S. P. Hau-Riege, S. Marchesini, B. W. Woods, S. Bajt, W. H. Benner, et al. 2006. Femtosecond diffractive imaging with a soft-X-ray free-electron laser. Nature Physics 2 (12):839–43. doi: 10.1038/nphys461.
- Chardin, H., K. Mercier, C. Frydman, and N. Vollmer. 2014. Surface plasmon resonance imaging: A method to measure the affinity of the antibodies in allergy diagnosis. Journal of Immunological Methods 405:23–8. doi: 10.1016/j.jim.2013.12.010.
- Chinthrajah, R. S., N. Purington, V. Sampath, S. Andorf, M. Manohar, M. Prunicki, X. Zhou, D. Tupa, and K. C. Nadeau. 2018. High dimensional immune biomarkers demonstrate differences in phenotypes and endotypes in food allergy and asthma. Annals of Allergy, Asthma and Immunology 121 (1):117–9.e1. doi: 10.1016/j.anai.2018.04.022.
- Cho, S.-W., D.-K. Kang, J.-B. Choo, A. J. Demllo, and S.-I. Chang. 2011. Recent advances in microfluidic technologies for biochemistry and molecular biology. BMB Reports 44 (11):705–12. doi: 10.5483/BMBRep.2011.44.11.705.
- Clement, C. C., A. Becerra, L. Yin, V. Zolla, L. Huang, S. Merlin, A. Follenzi, S. A. Shaffer, L. J. Stern, and L. Santambrogio. 2016. The dendritic cell major histocompatibility complex II (MHC II) peptidome derives from a variety of processing pathways and includes peptides with a broad spectrum of HLA-DM sensitivity. Journal of Biological Chemistry 291 (11):5576–95. doi: 10.1074/jbc.M115.655738.
- Codex Alimentarius Commission. 2001. Evaluation of allergenicity of genetically modified foods. Rome, Italy: Food and Agriculture Organization of the United Nations.
- Costa, J., S. L. Bavaro, S. Benedé, A. Diaz-Perales, C. Bueno-Diaz, E. Gelencser, J. Klueber, C. Larré, D. Lozano-Ojalvo, and R. Lupi. 2020. Are physicochemical properties shaping the allergenic potency of plant allergens? Clinical Reviews in Allergy and Immunology. doi: 10.1007/s12016-020-08810-9.
- Costa, J., C. Villa, K. Verhoeckx, T. Cirkovic-Velickovic, D. Schrama, P. Roncada, P. M. Rodrigues, C. Piras, L. Martín-Pedraza, and L. Monaci. 2021. Are physicochemical properties shaping the allergenic potency of animal allergens? Clinical Reviews in Allergy and Immunology. doi: 10.1007/s12016-020-08826-1.
- Couto, N., L. Davlyatova, C. A. Evans, and P. C. Wright. 2018. Application of the broadband collision-induced dissociation (bbCID) mass spectrometry approach for protein glycosylation and phosphorylation analysis. Rapid Communications in Mass Spectrometry 32 (2):75–85. doi: 10.1002/rcm.8016.
- Cox, K. M., S. P. Commins, B. J. Capaldo, L. J. Workman, T. A. E. Platts-Mills, E. D. Amir, J. A. Lannigan, A. J. Schuyler, and L. D. Erickson. 2019. An integrated framework using high-dimensional mass cytometry and fluorescent flow cytometry identifies discrete B cell subsets in patients with red meat allergy. Clinical & Experimental Allergy 49 (5):615–25. doi: 10.1111/cea.13322.
- Cressman, R. F., and G. Ladics. 2009. Further evaluation of the utility of "sliding window" FASTA in predicting cross-reactivity with allergenic proteins. Regulatory Toxicology and Pharmacology 54 (3):S20–S25. doi: 10.1016/j.yrtph.2008.11.006.
- Cretich, M., G. D. Carlo, C. Giudici, S. Pokoj, I. Lauer, S. Scheurer, and M. Chiari. 2009. Detection of allergen specific immunoglobulins by microarrays coupled to microfluidics. Proteomics 9 (8):2098–107. doi: 10.1002/pmic.200800651.
- Croote, D., S. Darmanis, K. C. Nadeau, and S. R. Quake. 2018. High-affinity allergen-specific human antibodies cloned from single IgE B cell transcriptomes. Science 362 (6420):1306–9. doi: 10.1126/science.aau2599.
- Cui, J., L. Y. Han, H. Li, C. Y. Ung, Z. Q. Tang, C. J. Zheng, Z. W. Cao, and Y. Z. Chen. 2007. Computer prediction of allergen proteins from sequence-derived protein structural and physicochemical properties. Molecular Immunology 44 (4):514–20. doi: 10.1016/j.molimm.2006.02.010.
- Dall’ Antonia, F., T. Pavkov-Keller, K. Zangger, and W. Keller. 2014. Structure of allergens and structure based epitope predictions. Methods 66 (1):3–21. doi: 10.1016/j.ymeth.2013.07.024.
- Dhondalay, G. K., E. Rael, S. Acharya, W. M. Zhang, V. Sampath, S. J. Galli, R. Tibshirani, S. D. Boyd, H. Maecker, K. C. Nadeau, et al. 2018. Food allergy and omics. Journal of Allergy and Clinical Immunology 141 (1):20–9. doi: 10.1016/j.jaci.2017.11.007.
- Emwas, A. H., R. Roy, R. T. McKay, L. Tenori, E. Saccenti, G. A. N. Gowda, D. Raftery, F. Alahmari, L. Jaremko, M. Jaremko, et al. 2019. NMR spectroscopy for metabolomics research. Metabolites 9 (7):123. doi: 10.3390/metabo9070.
- Fiers, M. W. E. J., G. A. Kleter, H. Nijland, A. A. C. M. Peijnenburg, J. P. Nap, and R. C. H. J. van Ham. 2004. Allermatch, a webtool for the prediction of potential allergenicity according to current FAO/WHO Codex alimentarius guidelines. BMC Bioinformatics 5 (1):133. doi: 10.1186/1471-2105-5-133.
- Franken, H., T. Mathieson, D. Childs, G. M. A. Sweetman, T. Werner, I. Tögel, C. Doce, S. Gade, M. Bantscheff, G. Drewes, et al. 2015. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nature Protocols 10 (10):1567–93. doi: 10.1038/nprot.2015.101.
- Gaetani, M., P. Sabatier, A. A. Saei, C. M. Beusch, Z. Yang, S. L. Lundstrom, and R. A. Zubarev. 2019. Proteome integral solubility alteration: A high-throughput proteomics assay for target deconvolution. Journal of Proteome Research 18 (11):4027–37. doi: 10.1021/acs.jproteome.9b00500.
- Gallat, F. X., N. Matsugaki, N. P. Coussens, K. J. Yagi, M. Boudes, T. Higashi, D. Tsuji, Y. Tatano, M. Suzuki, E. Mizohata, et al. 2014. In vivo crystallography at X-ray free-electron lasers: The next generation of structural biology. Philosophical Transactions of the Royal Society B: Biological Sciences 369 (1647):20130497. doi: 10.1098/rstb.2013.0497.
- Glab, T. K., and J. Boratynski. 2017. Potential of casein as a carrier for biologically active agents. Topics in Current Chemistry 375 (20). doi: 10.1007/s41061-017-0158-z.
- Goodman, R. E., M. Ebisawa, F. Ferreira, H. A. Sampson, R. van Ree, S. Vieths, J. L. Baumert, B. Bohle, S. Lalithambika, J. Wise, et al. 2016. AllergenOnline: A peer-reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Molecular Nutrition & Food Research 60 (5):1183–98. doi: 10.1002/mnfr.201500769.
- Goodman, R. E., S. Vieths, H. A. Sampson, D. Hill, M. Ebisawa, S. L. Taylor, and R. van Ree. 2008. Allergenicity assessment of genetically modified crops - what makes sense? Nature Biotechnology 26 (1):73–81. doi: 10.1038/nbt1343.
- Goswami, R., A. B. Blazquez, R. Kosoy, A. Rahman, A. Nowak-Węgrzyn, and M. C. Berin. 2017. Systemic innate immune activation in food protein-induced enterocolitis syndrome. Journal of Allergy and Clinical Immunology 139 (6):1885–96.e9. doi: 10.1016/j.jaci.2016.12.971.
- Guan, X., K. A. Noble, Y. Tao, K. H. Roux, S. K. Sathe, N. L. Young, and A. G. Marshall. 2015. Epitope mapping of 7S cashew antigen in complex with antibody by solution-phase H/D exchange monitored by FT-ICR mass spectrometry. Journal of Mass Spectrometry 50 (6):812–9. doi: 10.1002/jms.3589.
- Hamilton, R. G., S. S. Saini, and D. MacGlashan. 2012. Surface plasmon resonance analysis of free IgE in allergic patients receiving omalizumab (Xolair). Journal of Immunological Methods 383 (1-2):54–9. doi: 10.1016/j.jim.2012.05.015.
- Hiep, H. M., T. Endo, K. Kerman, M. Chikae, D. K. Kim, S. Yamamura, Y. Takamura, and E. Tamiya. 2007. A localized surface plasmon resonance based immunosensor for the detection of casein in milk. Science and Technology of Advanced Materials 8 (4):331–8. doi: 10.1016/j.stam.2006.12.010.
- Hirano, T., A. Koyanagi, K. Kotoshiba, Y. Shinkai, M. Kasai, T. Ando, A. Kaitani, K. Okumura, and J. Kitaura. 2018. The Fab fragment of anti-IgE Cε2 domain prevents allergic reactions through interacting with IgE-FcεRIα complex on rat mast cells. Scientific Reports 8 (1):14237. doi: 10.1038/s41598-018-32200-z.
- Hochwallner, H., U. Schulmeister, I. Swoboda, S. Spitzauer, and R. Valenta. 2014. Cow's milk allergy: From allergens to new forms of diagnosis, therapy and prevention. Methods 66 (1):22–33. doi: 10.1016/j.ymeth.2013.08.005.
- Holland, J. W., H. C. Deeth, and P. F. Alewood. 2004. Proteomic analysis of kappa-casein micro-heterogeneity. Proteomics 4 (3):743–52. doi: 10.1002/pmic.200300613.
- Holland, J. W., H. C. Deeth, and P. F. Alewood. 2006. Resolution and characterisation of multiple isoforms of bovine κ-casein by 2-DE following a reversible cysteine-tagging enrichment strategy. Proteomics 6 (10):3087–95. doi: 10.1002/pmic.200500780.
- Hummel, M., T. Wigger, and J. Brockmeyer. 2015. Characterization of mustard 2S albumin allergens by bottom-up, middle-down, and top-down proteomics: A consensus set of isoforms of Sin a 1. Journal of Proteome Research 14 (3):1547–56. doi: 10.1021/pr5012262.
- Inaba, I., H. Kuramitz, and K. Sugawara. 2016. Electrochemical sensing of casein based on the interaction between its phosphate groups and a ruthenium(III) complex. Analytical Sciences 32 (8):853–9. doi: 10.2116/analsci.32.853.
- Ivanciuc, O., C. H. Schein, and W. Braun. 2003. SDAP: Database and computational tools for allergenic proteins. Nucleic Acids Research 31 (1):359–62. doi: 10.1093/nar/gkg010.
- Jakobsen, C. G., U. Bodtger, L. K. Poulsen, and E. L. Roggen. 2005. Vaccination for birch pollen allergy: Comparison of the affinities of specific immunoglobulins E, G1 and G4 measured by surface plasmon resonance. Clinical & Experimental Allergy 35 (2):193–8. doi: 10.1111/j.1365-2222.2005.02160.x.
- Jemmerson, R., and Y. Paterson. 1986. Mapping epitopes on a protein antigen by the proteolysis of antigen-antibody complexes. Science 232 (4753):1001–4. doi: 10.1126/science.2422757.
- Joshi, A. A., M. W. Peczuh, C. V. Kumar, and J. F. Rusling. 2014. Ultrasensitive carbohydrate-peptide SPR imaging microarray for diagnosing IgE mediated peanut allergy. The Analyst 139 (22):5728–33. doi: 10.1039/C4AN01544D.
- Khoury, G. A., R. C. Baliban, and C. A. Floudas. 2011. Proteome-wide post-translational modification statistics: Frequency analysis and curation of the swiss-prot database. Scientific Reports 1 (1):90. doi: 10.1038/srep00090.
- King, C., R. S. Stein, J. L. Shamshina, and R. D. Rogers. 2017. Measuring the purity of chitin with a clean, quantitative solid-State NMR Method. ACS Sustainable Chemistry & Engineering 5 (9):8011–6. doi: 10.1021/acssuschemeng.7b01589.
- Koppelman, S. J., S. L. Hefle, S. L. Taylor, and G. A. H. de Jong. 2010. Digestion of peanut allergens Ara h 1, Ara h 2, Ara h 3, and Ara h 6: A comparative in vitro study and partial characterization of digestion-resistant peptides. Molecular Nutrition & Food Research 54 (12):1711–21. doi: 10.1002/mnfr.201000011.
- Kosoy, R., C. Agashe, A. Grishin, D. Y. Leung, R. A. Wood, S. H. Sicherer, S. M. Jones, A. W. Burks, W. F. Davidson, R. W. Lindblad, et al. 2016. Transcriptional profiling of egg allergy and relationship to disease phenotype. PLoS One 11 (10):e0163831. doi: 10.1371/journal.pone.0163831.
- Krushelnitsky, A., D. Reichert, and K. Saalwachter. 2013. Solid-state NMR approaches to internal dynamics of proteins: From picoseconds to microseconds and seconds. Accounts of Chemical Research 46 (9):2028–36. doi: 10.1021/ar300292p.
- Krutz, N. L., J. Winget, C. A. Ryan, R. Wimalasena, S. Maurer-Stroh, R. J. Dearman, I. Kimber, and G. F. Gerberick. 2019. Proteomic and bioinformatic analyses for the identification of proteins with low allergenic potential for hazard assessment. Toxicological Sciences 170 (1):210–22. doi: 10.1093/toxsci/kfz078.
- Ladizhansky, V. 2019. Nuclear magnetic resonance spectroscopy. Solid-state NMR of macromolecules. In Encyclopedia of analytical science, ed. P. Worsfold, C. Poole, A. Townshend, and M. Miró, 414–26. Amsterdam, Netherlands: Elsevier Inc. doi: 10.1016/B978-0-12-409547-2.14083-1.
- Laffer, S., C. Lupinek, I. Rauter, M. Kneidinger, A. Drescher, J. H. Jordan, M. T. Krauth, P. Valent, F. Kricek, S. Spitzauer, et al. 2008. A high-affinity monoclonal anti-IgE antibody for depletion of IgE and IgE-bearing cells. Allergy 63 (6):695–702. doi: 10.1111/j.1398-9995.2008.01664.x.
- Lanucara, F., and C. E. Eyers. 2013. Top-down mass spectrometry for the analysis of combinatorial post-translational modifications. Mass Spectrometry Reviews 32 (1):27–42. doi: 10.1002/mas.21348.
- Larocca, M., G. Martelli, G. Grossi, M. C. Padula, P. Riccio, and R. Rossano. 2013. Peel LTP (Pru p 3)-the major allergen of peach-is methylated. A proteomic study. Food Chemistry 141 (3):2765–71. doi: 10.1016/j.foodchem.2013.04.082.
- Lauer, I., K. Foetisch, D. Kolarich, B. K. Ballmer-Weber, A. Conti, F. Altmann, S. Vieths, and S. Scheurer. 2004. Hazelnut (Corylus avellana) vicilin Cor a 11: Molecular characterization of a glycoprotein and its allergenic activity. Biochemical Journal 383 (2):327–34. doi: 10.1042/BJ20041062.
- Le, T. T., H. C. Deeth, and L. B. Larsen. 2017. Proteomics of major bovine milk proteins: Novel insights. International Dairy Journal 67:2–15. doi: 10.1016/j.idairyj.2016.11.016.
- Legler, J., D. Zalko, F. Jourdan, M. Jacobs, B. Fromenty, P. Balaguer, W. Bourguet, V. Munic Kos, A. Nadal, C. Beausoleil, et al. 2020. The GOLIATH project: Towards an internationally harmonised approach for testing metabolism disrupting compounds. International Journal of Molecular Sciences 21 (10):3480. doi: 10.3390/ijms21103480.
- Leitner, A. 2016. Cross-linking and other structural proteomics techniques: How chemistry is enabling mass spectrometry applications in structural biology. Chemical Science 7 (8):4792–803. doi: 10.1039/C5SC04196A.
- Leitner, A., M. Faini, F. Stengel, and R. Aebersold. 2016. Crosslinking and mass spectrometry: An integrated technology to understand the structure and function of molecular machines. Trends in Biochemical Sciences 41 (1):20–32. doi: 10.1016/j.tibs.2015.10.008.
- Li, J. X., K. Shefcheck, J. Callahan, and C. Fenselau. 2009. Primary sequence and site-selective hydroxylation of prolines in isoforms of a major peanut allergen protein Ara h 2. Protein Science 19:174–82. doi: 10.1002/pro.295.
- Li, L., C. Wang, S. Qiang, J. Zhao, S. Song, W. Jin, B. Wang, Y. Zhang, L. Huang, and Z. Wang. 2016. Mass spectrometric analysis of N-glycoforms of soybean allergenic glycoproteins separated by SDS-PAGE. Journal of Agricultural and Food Chemistry 64 (39):7367–76. doi: 10.1021/acs.jafc.6b02773.
- Lin, H. Y., C. H. Huang, J. Park, D. Pathania, C. M. Castro, A. Fasano, R. Weissleder, and H. Lee. 2017. Integrated magneto-chemical sensor for on-site food allergen detection. ACS Nano 11 (10):10062–9. doi: 10.1021/acsnano.7b04318.
- López-Fandiño, R. 2020. Role of dietary lipids in food allergy. Critical Reviews in Food Science and Nutrition 60 (11):1797–814. doi: 10.1080/10408398.2019.1602025.
- Loquet, A., N. E. Mammeri, J. Stanek, M. Berbon, B. Bardiaux, G. Pintacuda, and B. Habenstein. 2018. 3D structure determination of amyloid fibrils using solid-state NMR spectroscopy. Methods 138-139:26–38. doi: 10.1016/j.ymeth.2018.03.014.
- Lozano-Ojalvo, D., S. Benedé, and J. van Bilsen. 2021. Food allergy: Etiology, allergens, and analytical strategies. In Reference module in food science, ed. A. Cifuentes, 175–196. Amsterdam, Netherlands: Elsevier. doi: 10.1016/B978-0-08-100596-5.22845-X.
- Lu, Y., T. Ohshima, and H. Ushio. 2004. Rapid detection of fish major allergen parvalbumin by surface plasmon resonance biosensor. Journal of Food Science 69 (8):C652–8. doi: 10.1111/j.1750-3841.2004.tb18013.x.
- Maleki, S. J., S. Y. Chung, E. T. Champagne, and J. P. Raufman. 2000. The effects of roasting on the allergenic properties of peanut proteins. Journal of Allergy and Clinical Immunology 106 (4):763–8. doi: 10.1067/mai.2000.109620.
- Martinez Molina, D., R. Jafari, M. Ignatushchenko, T. Seki, E. A. Larsson, C. Dan, L. Sreekumar, Y. Cao, and P. Nordlund. 2013. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341 (6141):84–7. doi: 10.1126/science.1233606.
- Matsuo, H., T. Yokooji, and T. Taogoshi. 2015. Common food allergens and their IgE-binding epitopes. Allergology International 64 (4):332–43. doi: 10.1016/j.alit.2015.06.009.
- Maurer-Stroh, S., N. L. Krutz, P. S. Kern, V. Gunalan, M. N. Nguyen, V. Limviphuvadh, F. Eisenhaber, and G. F. Gerberick. 2019. AllerCatPro - Prediction of protein allergenicity potential from the protein sequence. Bioinformatics 35 (17):3020–7. doi: 10.1093/bioinformatics/btz029.
- Mazzucchelli, G., T. Holzhauser, T. Cirkovic Velickovic, A. Diaz-Perales, E. Molina, P. Roncada, P. Rodrigues, K. Verhoeckx, and K. Hoffmann-Sommergruber. 2018. Current (food) allergenic risk assessment: Is it fit for novel foods? Status Quo and identification of gaps. Molecular Nutrition & Food Research 62 (1):1700278. doi: 10.1002/mnfr.201700278.
- Michel, D., F. Xiao, and K. Alameh. 2017. A compact, flexible fiber-optic surface plasmon resonance sensor with changeable sensor chips. Sensors and Actuators B: Chemical 246:258–61. doi: 10.1016/j.snb.2017.02.064.
- Moffat, J. G., F. Vincent, J. A. Lee, J. Eder, and M. Prunotto. 2017. Opportunities and challenges in phenotypic drug discovery: An industry perspective. Nature Reviews Drug Discovery 16 (8):531–43. doi: 10.1038/nrd.2017.111.
- Montiel, R. V. R., S. Campuzano, R. M. Torrente-Rodriguez, A. J. Reviejo, and J. M. Pingarron. 2016. Electrochemical magnetic beads-based immunosensing platform for the determination of alpha-lactalbumin in milk. Food Chemistry 213:595–601. doi: 10.1016/j.foodchem.2016.07.004.
- Montiel, V. R. V., S. Campuzano, F. Conzuelo, R. M. Torrente-Rodriguez, M. Gamella, A. J. Reviejo, and J. M. Pingarron. 2015. Electrochemical magnetoimmunosensing platform for determination of the milk allergen beta-lactoglobulin. Talanta 131:156–62. doi: 10.1016/j.talanta.2014.07.076.
- Montiel, V. R. V., A. Pellicano, S. Campuzano, R. M. Torrente-Rodriguez, A. J. Reviejo, M. S. Cosio, and J. M. Pingarron. 2016. Electrochemical detection of peanuts at trace levels in foods using a magnetoimmunosensor for the allergenic protein Ara h 2. Sensors and Actuators B: Chemical 236:825–33. doi: 10.1016/j.snb.2016.01.123.
- Montiel, V. R. V., R. M. Torrente-Rodriguez, G. G. de Rivera, A. J. Reviejo, C. Cuadrado, R. Linacero, F. J. Gallego, S. Campuzano, and J. M. Pingarron. 2017. Amperometric determination of hazelnut traces by means of Express PCR coupled to magnetic beads assembled on disposable DNA sensing scaffolds. Sensors and Actuators B: Chemical 245:895–902. doi: 10.1016/j.snb.2017.02.041.
- Mueller, G. A. 2017. Contributions and future directions for structural biology in the study of allergens. International Archives of Allergy and Immunology 174 (2):57–66. doi: 10.1159/000481078.
- Muh, H. C., J. C. Tong, and M. T. Tammi. 2009. AllerHunter: A SVM-pairwise system for assessment of allergenicity and allergic cross-reactivity in proteins. PLoS One 4 (6):e5861. doi: 10.1371/journal.pone.0005861.
- Mukai, K., N. Gaudenzio, S. Gupta, N. Vivanco, S. C. Bendall, H. T. Maecker, R. S. Chinthrajah, M. Tsai, K. C. Nadeau, and S. J. Galli. 2017. Assessing basophil activation by using flow cytometry and mass cytometry in blood stored 24 hours before analysis. Journal of Allergy and Clinical Immunology 139 (3):889–99.e11. doi: 10.1016/j.jaci.2016.04.060.
- Neutze, R., R. Wouts, D. van der Spoel, E. Weckert, and J. Hajdu. 2000. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406 (6797):752–7. doi: 10.1038/35021099.
- Nugraha, R., S. D. Kamath, E. Johnston, K. R. Zenger, J. M. Rolland, R. E. O'Hehir, and A. L. Lopata. 2018. Rapid and comprehensive discovery of unreported shellfish allergens using large-scale transcriptomic and proteomic resources. Journal of Allergy and Clinical Immunology 141 (4):1501–4.e8. doi: 10.1016/j.jaci.2017.11.028.
- Ocana, C., A. Hayat, R. K. Mishra, A. Vasilescu, M. del Valle, and J. L. Marty. 2015. Label free aptasensor for Lysozyme detection: A comparison of the analytical performance of two aptamers. Bioelectrochemistry 105:72–7. doi: 10.1016/j.bioelechem.2015.05.009.
- Olsen, J. V., and M. Mann. 2013. Status of large-scale analysis of post-translational modifications by mass spectrometry. Molecular & Cellular Proteomics 12 (12):3444–52. doi: 10.1074/mcp.O113.034181.
- Pandeswari, P. B., and V. Sabareesh. 2019. Middle-down approach: A choice to sequence and characterize proteins/proteomes by mass spectrometry. RSC Advances 9 (1):313–44. doi: 10.1039/C8RA07200K.
- Pascovici, D., J. X. Wu, M. J. McKay, C. Joseph, Z. Noor, K. Kamath, Y. Wu, S. Ranganathan, V. Gupta, and M. Mirzaei. 2018. Clinically relevant post-translational modification analyses—Maturing workflows and bioinformatics tools. International Journal of Molecular Sciences 20 (1):16. doi: 10.3390/ijms20010016.
- Pekar, J., D. Ret, and E. Untersmayr. 2018. Stability of allergens. Molecular Immunology 100:14–20. doi: 10.1016/j.molimm.2018.03.017.
- Pereira-Barros, M. A., M. F. Barroso, L. Martin-Pedraza, E. Vargas, S. Benede, M. Villalba, J. M. Rocha, S. Campuzano, and J. M. Pingarron. 2019. Direct PCR-free electrochemical biosensing of plant-food derived nucleic acids in genomic DNA extracts. Application to the determination of the key allergen Sola l 7 in tomato seeds. Biosensors and Bioelectronics 137:171–7. doi: 10.1016/j.bios.2019.05.011.
- Piazza, I., K. Kochanowski, V. Cappelletti, T. Fuhrer, E. Noor, U. Sauer, and P. Picotti. 2018. A map of protein-metabolite interactions reveals principles of chemical communication. Cell 172 (1-2):358–72.e23. doi: 10.1016/j.cell.2017.12.006.
- Pilolli, R., A. Visconti, and L. Monaci. 2015. Rapid and label-free detection of egg allergen traces in wines by surface plasmon resonance biosensor. Analytical and Bioanalytical Chemistry 407 (13):3787–97. doi: 10.1007/s00216-015-8607-4.
- Pol, E., R. Karlsson, H. Roos, A. Jansson, B. Z. Xu, A. Larsson, T. Jarhede, G. Franklin, A. Fuentes, and S. Persson. 2007. Biosensor-based characterization of serum antibodies during development of an anti-IgE immunotherapeutic against allergy and asthma. Journal of Molecular Recognition 20 (1):22–31. doi: 10.1002/jmr.804.
- Rahman, A. M. A., A. L. Lopata, R. E. O'Hehir, J. J. Robinson, J. H. Banoub, and R. J. Helleur. 2010. Characterization and de novo sequencing of snow crab tropomyosin enzymatic peptides by both electrospary ionization and matrix-assisted laser desorption ionization QqToF tandem mass spectrometry. Journal of Mass Spectrometry 45:372–81. doi: 10.1002/jms.1721.
- Ramesh, K. R., R. Hemalatha, C. A. Vijayendra, U. Z. S. Arshi, S. B. Dushyant, and K. B. Dinesh. 2016. Transcriptome analysis of Solanum melongena L. (eggplant) fruit to identify putative allergens and their epitopes. Gene 576 (1):64–71. doi: 10.1016/j.gene.2015.09.064.
- Ravikiran, B., and R. Mahalakshmi. 2014. Unusual post-translational protein modifications: The benefits of sophistication. RSC Advances 4 (64):33958–74. doi: 10.1039/C4RA04694C.
- Ray, S., G. Mehta, and S. Srivastava. 2010. Label-free detection techniques for protein microarrays: Prospects, merits and challenges. Proteomics 10 (4):731–48. doi: 10.1002/pmic.200900458.
- Remington, B., H. C. H. Broekman, W. M. Blom, A. Capt, R. W. R. Crevel, I. Dimitrov, C. K. Faeste, R. Fernandez-Canton, S. Giavi, G. F. Houben, et al. 2018. Approaches to assess IgE mediated allergy risks (sensitization and cross-reactivity) from new or modified dietary proteins. Food and Chemical Toxicology 112:97–107. doi: 10.1016/j.fct.2017.12.025.
- Sathe, S. K., S. S. Teuber, and K. H. Roux. 2005. Effects of food processing on the stability of food allergens. Biotechnology Advances 23 (6):423–9. doi: 10.1016/j.biotechadv.2005.05.008.
- Savitski, M. M., F. B. M. Reinhard, H. Franken, T. Werner, M. F. Savitski, D. Eberhard, D. M. Molina, R. Jafari, R. B. Dovega, S. Klaeger, et al. 2014. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science 346 (6205):1255784. doi: 10.1126/science.1255784.
- Savitski, M. M., N. Zinn, M. Faelth-Savitski, D. Poeckel, S. Gade, I. Becher, M. Muelbaier, A. J. Wagner, K. Strohmer, T. Werner, et al. 2018. Multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis. Cell 173 (1):260–74.e25. doi: 10.1016/j.cell.2018.02.030.
- Schmidt, C., and H. Urlaub. 2017. Combining cryo-electron microscopy (cryo-EM) and cross-linking mass spectrometry (CX-MS) for structural elucidation of large protein assemblies. Current Opinion in Structural Biology 46:157–68. doi: 10.1016/j.sbi.2017.10.005.
- Schopper, S., A. Kahraman, P. Leuenberger, Y. Feng, I. Piazza, O. Müller, P. J. Boersema, and P. Picotti. 2017. Measuring protein structural changes on a proteome-wide scale using limited proteolysis-coupled mass spectrometry. Nature Protocols 12 (11):2391–410. doi: 10.1038/nprot.2017.100.
- Silvanovich, A., M. A. Nemeth, P. Song, R. Herman, L. Tagliani, and G. A. Bannon. 2006. The value of short amino acid sequence matches for prediction of protein allergenicity. Toxicological Sciences 90 (1):252–8. doi: 10.1093/toxsci/kfj068.
- Simoni, Y., M. H. Y. Chng, S. Li, M. Fehlings, and E. W. Newell. 2018. Mass cytometry: a powerful tool for dissecting the immune landscape. Current Opinion in Immunology 51:187–96. doi: 10.1016/j.coi.2018.03.023.
- Singh, R., P. P. Sharma, R. E. Baltus, and I. I. Suni. 2010. Nanopore immunosensor for peanut protein Ara h1. Sensors and Actuators B: Chemical 145 (1):98–103. doi: 10.1016/j.snb.2009.11.039.
- Sipova, H., and J. Homola. 2013. Surface plasmon resonance sensing of nucleic acids: A review. Analytica Chimica Acta 773:9–23. doi: 10.1016/j.aca.2012.12.040.
- Sirvent, S., Palomares, O. A. Vereda, M. Villalba, J. Cuesta, ‐Herranz, and R. Rodríguez. 2009. nsLTP and profilin are allergens in mustard seeds: Cloning, sequencing and recombinant production of Sin a 3 and Sin a 4. Clinical & Experimental Allergy 39 (12):1929–36. doi: 10.1111/j.1365-2222.2009.03382.x.
- Smole, U., M. Bublin, C. Radauer, C. Ebner, and H. Breiteneder. 2008. Mal d 2, the thaumatin-like allergen from apple, is highly resistant to gastrointestinal digestion and thermal processing. International Archives of Allergy and Immunology 147 (4):289–98. doi: 10.1159/000144036.
- Spitzer, M. H., and G. P. Nolan. 2016. Mass cytometry: Single cells, many features. Cell 165 (4):780–91. doi: 10.1016/j.cell.2016.04.019.
- Stadler, M. B., and B. M. Stadler. 2003. Allergenicity prediction by protein sequence. The FASEB Journal 17 (9):1141–3. doi: 10.1096/fj.02-1052fje.
- Stoeckius, M., C. Hafemeister, W. Stephenson, B. Houck-Loomis, P. K. Chattopadhyay, H. Swerdlow, R. Satija, and P. Smibert. 2017. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods 14 (9):865–8. doi: 10.1038/nmeth.4380.
- Sugahara, M., E. Mizohata, E. Nango, M. Suzuki, T. Tanaka, T. Masuda, R. Tanaka, T. Shimamura, Y. Tanaka, C. Suno, et al. 2015. Grease matrix as a versatile carrier of proteins for serial crystallography. Nature Methods 12 (1):61–3. doi: 10.1038/nmeth.3172.
- Sun, S., Y. Han, S. Paramasivam, S. Yan, A. E. Siglin, J. C. Williams, I. J. L. Byeon, J. Ahn, A. M. Gronenborn, and T. Polenova. 2012. Solid-state NMR spectroscopy of protein complexes. Methods in Molecular Biology 831:303–31. doi: 10.1007/978-1-61779-480-3_17.
- Syahir, A., K. Usui, K.-Y. Tomizaki, K. Kajikawa, and H. Mihara. 2015. Label and label-free detection techniques for protein microarrays. Microarrays 4 (2):228–44. doi: 10.3390/microarrays4020228.
- Tao, B., K. Bernardo, P. Eldi, N. Chegeni, M. Wiese, A. Colella, A. Kral, J. Hayball, W. Smith, K. Forsyth, et al. 2016. Extended boiling of peanut progressively reduces IgE allergenicity while retaining T cell reactivity. Clinical & Experimental Allergy 46 (7):1004–14. doi: 10.1111/cea.12740.
- Thomas, K., C. Herouet-Guicheney, G. Ladics, G. Bannon, A. Cockburn, R. Crevel, J. Fitzpatrick, C. Mills, L. Privalle, and S. Vieths. 2007. Evaluating the effect of food processing on the potential human allergeni city of novel proteins: International workshop report. Food and Chemical Toxicology 45 (7):1116–22. doi: 10.1016/j.fct.2006.12.016.
- Tordesillas, L., N. Cubells-Baeza, C. Gomez-Casado, C. Berin, V. Esteban, W. Barcik, L. O’Mahony, C. Ramirez, L. F. Pacios, M. Garrido-Arandia, et al. 2017. Mechanisms underlying induction of allergic sensitization by Pru p 3. Clinical & Experimental Allergy 47 (11):1398–408. doi: 10.1111/cea.12962.
- Tordesillas, L., A. H. Rahman, H. A. Sampson, and M. C. Berin. 2016. Profiling the immune response to peanut using mass cytometry. Journal of Allergy and Clinical Immunology 137 (2):AB74. doi: 10.1016/j.jaci.2015.12.251.
- Trejo, R., T. Dokland, J. Jurat-Fuentes, and F. Harte. 2011. Cryo-transmission electron tomography of native casein micelles from bovine milk. Journal of Dairy Science 94 (12):5770–5. doi: 10.3168/jds.2011-4368.
- Turkowsky, D., P. Lohmann, M. Muhlenbrink, T. Schubert, L. Adrian, T. Goris, N. Jehmlich, and M. von Bergen. 2019. Thermal proteome profiling allows quantitative assessment of interactions between tetrachloroethene reductive dehalogenase and trichloroethene. Journal of Proteomics 192:10–7. doi: 10.1016/j.jprot.2018.05.018.
- Upton, J., and A. Nowak-Wegrzyn. 2018. The impact of baked egg and baked milk diets on IgE- and non-IgE-mediated allergy. Clinical Reviews in Allergy & Immunology 55 (2):118–38. doi: 10.1007/s12016-018-8669-0.
- van der Wel, P. C. A. 2017. Insights into protein misfolding and aggregation enabled by solid-state NMR spectroscopy. Solid State Nuclear Magnetic Resonance 88:1–14. doi: 10.1016/j.ssnmr.2017.10.001.
- Vedadi, M., F. H. Niesen, A. Allali-Hassani, O. Y. Fedorov, P. J. Finerty, G. A. Wasney, R. Yeung, C. Arrowsmith, L. J. Ball, H. Berglund, et al. 2006. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proceedings of the National Academy of Sciences 103 (43):15835–40. doi: 10.1073/pnas.0605224103.
- Verhoeckx, K., H. Broekman, A. Knulst, and G. Houben. 2016. Allergenicity assessment strategy for novel food proteins and protein sources. Regulatory Toxicology and Pharmacology 79:118–24. doi: 10.1016/j.yrtph.2016.03.016.
- Vickery, B. P., S. Chin, and A. W. Burks. 2011. Pathophysiology of food allergy. Pediatric Clinics of North America 58 (2):363–76. doi: 10.1016/j.pcl.2011.02.012.
- Villas-Boas, M. B., S. Benedé, R. de Lima Zollner, F. M. Netto, and E. Molina. 2015. Epitopes resistance to the simulated gastrointestinal digestion of β-lactoglobulin submitted to two-step enzymatic modification. Food Research International 72:191–7. doi: 10.1016/j.foodres.2015.03.044.
- Wang, J., Y. B. Yu, Y. A. Zhao, D. B. Zhang, and J. Li. 2013. Evaluation and integration of existing methods for computational prediction of allergens. BMC Bioinformatics 14 (Suppl 4):S1. doi: 10.1186/1471-2105-14-S4-S1.
- Wang, J., D. B. Zhang, and J. Li. 2013. PREAL: Prediction of allergenic protein by maximum Relevance Minimum Redundancy (mRMR) feature selection. BMC Systems Biology 7 (Suppl 5):S9. doi: 10.1186/1752-0509-7-S5-S9.
- Wang, X. X., X. W. Zhao, D. W. Huang, X. C. Pan, Y. X. Qi, Y. X. Yang, H. L. Zhao, and G. L. Cheng. 2017. Proteomic analysis and cross species comparison of casein fractions from the milk of dairy animals. Scientific Reports 7 (1):9. doi: 10.1038/srep43020.
- Watson, C. T., A. T. Cohain, R. S. Griffin, Y. Chun, A. Grishin, H. Hacyznska, G. E. Hoffman, N. D. Beckmann, H. Shah, P. Dawson, et al. 2017. Integrative transcriptomic analysis reveals key drivers of acute peanut allergic reactions. Nature Communications 8 (1):13. doi: 10.1038/s41467-017-02188-7.
- Weng, X., G. Gaur, and S. Neethirajan. 2016. Rapid detection of food allergens by microfluidics ELISA-based optical sensor. Biosensors 6 (2):24. 10. doi: 10.3390/bios60200.
- Willison, L. N., P. Tripathi, G. Sharma, S. S. Teuber, S. K. Sathe, and K. H. Roux. 2011. Cloning, expression and patient IgE reactivity of recombinant Pru du 6, an 11S globulin from almond. International Archives of Allergy and Immunology 156 (3):267–81. doi: 10.1159/000323887.
- Willison, L. N., Q. Zhang, M. Su, S. S. Teuber, S. K. Sathe, and K. H. Roux. 2013. Conformational epitope mapping of Pru du 6, a major allergen from almond nut. Molecular Immunology 55 (3-4):253–63. doi: 10.1016/j.molimm.2013.02.004.
- Wu, X. L., Y. Li, B. Liu, Y. Feng, W. Y. He, Z. G. Liu, L. Z. Liu, Z. M. Wang, and H. Z. Huang. 2016. Two-site antibody immunoanalytical detection of food allergens by surface plasmon resonance. Food Analytical Methods 9 (3):582–8. doi: 10.1007/s12161-015-0232-5.
- Wylie, B. J., H. Q. Do, C. G. Borcik, E. P. Hardy, and P. C. der Wel. 2016. Advances in solid-state NMR of membrane proteins. Molecular Physics 114 (24):3598–09. doi: 10.1080/00268976.2016.1252470.
- Yakubu, R. R., E. Nieves, and L. M. Weiss. 2019. The methods employed in mass spectrometric analysis of posttranslational modifications (PTMs) and protein–protein interactions (PPIs). In Advancements of mass spectrometry in biomedical research, ed. A. G. Woods and C. C. Darie, 169–98. Cham: Springer International Publishing. doi: 10.1007/978-3-030-15950-4_10.
- Yan, Y., G. Chen, H. Wei, R. Y.-C. Huang, J. Mo, D. L. Rempel, A. A. Tymiak, and M. L. Gross. 2014. Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. Journal of the American Society for Mass Spectrometry 25 (12):2084–92. doi: 10.1007/s13361-014-0993-x.
- Yanase, Y., T. Hiragun, K. Ishii, T. Kawaguchi, T. Yanase, M. Kawai, K. Sakamoto, and M. Hide. 2014. Surface plasmon resonance for cell-based clinical diagnosis. Sensors 14 (3):4948–59. doi: 10.3390/s140304948.
- Yanase, Y., T. Hiragun, T. Yanase, T. Kawaguchi, K. Ishii, and M. Hide. 2012. Evaluation of peripheral blood basophil activation by means of surface plasmon resonance imaging. Biosensors and Bioelectronics 32 (1):62–8. doi: 10.1016/j.bios.2011.11.023.
- Zhang, Q., K. A. Noble, Y. Mao, N. L. Young, S. K. Sathe, K. H. Roux, and A. G. Marshall. 2013. Rapid screening for potential epitopes reactive with a polycolonal antibody by solution-phase H/D exchange monitored by FT-ICR mass spectrometry. Journal of the American Society for Mass Spectrometry 24 (7):1016–25. doi: 10.1007/s13361-013-0644-7.
- Zhou, J. R., Q. Q. Qi, C. Wang, Y. F. Qian, G. M. Liu, Y. B. Wang, and L. L. Fu. 2019. Surface plasmon resonance (SPR) biosensors for food allergen detection in food matrices. Biosensors and Bioelectronics 142:111449. doi: 10.1016/j.bios.2019.
- Zhou, J. R., Y. B. Wang, Y. F. Qian, T. Zhang, L. Zheng, and L. L. Fu. 2020. Quantification of shellfish major allergen tropomyosin by SPR biosensor with gold patterned Biochips. Food Control 107:106547. doi: 10.1016/j.foodcont.2019.02.041.
- Ziegler, S., V. Pries, C. Hedberg, and H. Waldmann. 2013. Target identification for small bioactive molecules: Finding the needle in the haystack. Angewandte Chemie International Edition 52 (10):2744–92. doi: 10.1002/anie.201208749.
