?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Reconstituted high density lipoprotein (rHDL) has been regarded as a promising brain-targeting vehicle for anti-glioma drugs under the mediation of apolipoprotein A-I (apoA-I). However, some stability issues relating to drug leakage and consequent reduced targeting efficiency in the course of discoidal rHDL (d-rHDL) circulating in blood hinder its broad application. The objective of the study was to develop a novel stabilized d-rHDL by replacing cholesterol and apoA-I with mono-cholesterol glutarate (MCG) modified apoA-I (termed as mA) and to evaluate its allosteric behavior and glioma targeting. MCG was synthesized through esterifying the hydroxyl of cholesterol with glutaric anhydride and characterized by FI-IR and 1H NMR. d-rHDL assembled with mA (termed as m-d-rHDL) presented similar properties such as minute particle size and disk-like appearance resembling nascent HDL. Morphological transformation observation and in vitro release plots convinced that the modification of cholesterol could effectively inhibit the remolding of d-rHDL. The uptake of m-d-rHDL by LCAT-pretreated bEND.3 cells was significantly higher than that of d-rHDL, thereby serving as another proof for the capability of m-d-rHDL in enhancing targeting property. Besides, apoA-I anchoring into m-d-rHDL played a critical role in the endocytosis process into bEND.3 cells and C6 cells, which implied the possibility of traversing blood brain barrier and accumulating in the brain and glioma. These results suggested that the modification toward cholesterol to improve the stability of d-rHDL is advantageous, and that this obtained m-d-rHDL revealed great potential for realization of suppressing the remolding of d-rHDL in the brain-targeted treatment of glioma for drug delivery.
1. Introduction
Astrocytoma is the most common type of gliom accounting for about 30% of brain tumors and about 65% of glioblastomas (Yang et al., Citation2020). Surgical excision is regarded as the major treatment supplemented by radiotherapy and chemotherapy, which is still confronted with unclear removal of all the tumor tissues or sustained impairment from radiotherapy and chemotherapy to normal organs (Lavrador et al., Citation2017). Increasing numbers of drugs are confronted with the problems of crossing the blood–brain barrier (BBB) even though they have been proved to possess the ability to inhibit the proliferation of glioma cells in vitro (Chen et al., Citation2016; Lin et al., Citation2017; Zhu et al., Citation2017; Qian et al., Citation2018). However, the terrible incurability and poor prognosis of astroglioma are precisely ascribed to the existing sophisticated physiological structure of brain and the difficulties in breaching BBB (Brandner and Jaunmuktane, Citation2018). Recently, one of the novel brain-targeting drug delivery system, lipoprotein-based nanoparticle, has attracted the attention for their unique features to improve the problems mentioned above (Cui et al., Citation2018).
High density lipoprotein (HDL), one of endogenous human plasma lipoproteins, has been successfully used to deliver a large number of lipophilic drugs due to its excellent characteristics such as structural features and biocompatibility (Nicholls et al., Citation2018). Natural HDL has a Stokes diameter of 5–17 nm and exists in two distinct forms: the spherical particle has a hydrophobic core of triglyceride (TG) and cholesterol ester (CE) surrounding with a phospholipid monolayer in which apolipoproteins (apos) are embedded and the other discoidal shape composes of only apos and phospholipid bilayer (phospholipid and cholesterol) (Lund-Katz et al., Citation2003; Tosheska Trajkovska and Topuzovska, Citation2017; Cooke et al., Citation2018). Apolipoprotein A-I (apoA-I), accounting for approximately 70% of the HDL protein mass, plays a vital role in assisting the formation and stabilization of HDL, as well as generating endocytosis via scavenger receptor B type I (SR-BI) mediated lipid metabolism (Vaisar et al., Citation2007). In the past decades, reconstituted HDL (rHDL) with physicochemical properties similar to those of native HDL has been well established as a targeting delivery vehicle (Marsche, Citation2015). These HDL-like nanoparticles offer attractive attributes such as favorable structure, endogenesis, target effect, a long circulation time in plasma and capacity to escape from recognition and elimination by reticuloendothelial system (RES) (Gordts et al., Citation2013). Recently, there is growing evidence showing the brain targeting effect of rHDL for cerebral disease (Song et al., Citation2014). Petri and Kratzer revealed that apoA-I could bind with SR-BI specifically expressed on brain microvascular endothelial cells and astrocytes, which contributes to transferring HDL into brain to mediate intracerebral lipid metabolism in the form of transcellular transport of endothelial cells and uptake of astrocytes (Kratzer et al., Citation2007; Petri et al., Citation2007). Besides, it has also been reported that the liposomes decorated with apoA-I could be uptaken by rat glioma cells (C6 cells) via SR-BI which is also abundant on the surface of cancer cells (Hong et al., Citation2007; Park et al., Citation2010; Lu et al., Citation2015; Rajora and Zheng, Citation2016). Relating to or produced by the mediated effect of apoA-I as above, rHDL might be endowed with the excellent capacity of transporting across BBB, accumulating in the brain and tumor.
Unlike spherical rHDL transporting CE into tumor cells, discoidal rHDL (d-rHDL) has been proved suitable to simulate the discoidal nascent HDL in terms of mediating tumor intracellular cholesterol efflux and destroying the lipid raft structure of cancer cells to accelerate necrosis and apoptosis of tumor cells (Tsompanidi et al., Citation2010), exerting synergistic effect on the basis of anti-tumor drugs. Besides, d-rHDL has a much stronger affinity with SR-BI, which is about an order of magnitude higher than spherical rHDL (Zhang, Citation2004). Undoubtedly, it is more efficient for d-rHDL to achieve active drug transport into tumor cells. Coupled with the unique liposome-like structure, d-rHDL possesses the biomimetic property of cell membrane fusion and is more likely to realize intracellular delivery of drugs (Pedersbaek et al., Citation2019). Based on the above considerations, d-rHDL seems to have the superiority as a promising brain-targeted vehicle for anti-glioma drugs.
However, amphipathic cholesterol in the primary d-HDL could be esterified to generate hydrophobic CE with the activation of lecithin-cholesterol acyltransferase (LCAT) in blood, leading to the structure conversion from nascent d-HDL to mature spherical HDL, which alleged the reconstruction of HDL (Florence and Halbert, Citation1991). Serving as drug delivery system, d-rHDL may also go through the same allosteric process in blood circulation, which would cause numerous leakage of core-enveloped drug and the attendant sharp decrease in targeting efficiency induced by LCAT during the targeted-transport process to the brain (Wang et al., Citation2013). ApoA-I, phospholipid, and cholesterol are regarded as essential factors in the remodeling of d-rHDL. Once apoA-I is activated, LCAT would transfer sn-2 acyl group of phospholipid to hydroxyl group of cholesterol to generate remodeling process (Carlucci et al., Citation2012). Any modifications relative to the above three factors might terminate the remodeling induced by LCAT (Nobecourt et al., Citation2007). With respect to the heterogeneity of phospholipids, it might be more reasonable to esterify the hydroxyl of cholesterol with glutaric anhydride to get mono-cholesterol glutarate (MCG), eventually erasing the enzymatic substrate and escaping the catalytic attack of LCAT.
In this study, d-rHDL was constructed to mimic the structure and function of lipoprotein and realize the effect of astrocytoma targeting. MCG was identified by FI-IR and 1H NMR, and then covalently attached with apoA-I to synthesize MCG modified apoA-I (termed as mA). The resultant mA was involved in the formation of stable rHDL (termed as m-d-rHDL). The m-d-rHDL was characterized by size distribution, zeta potential, transmission electron microscopy (TEM). The colloidal stability and allosteric behavior including microscope observation and coumarin 6 release of d-rHDL and m-d-rHDL were examined and compared in vitro. Uptakes of m-d-rHDL by bEND.3 cells and C6 cells were utilized to evaluate the ability to cross BBB and target glioma. Cytotoxicity study was also evaluated using bEND.3 cell line and C6 cell line.
2. Materials and methods
2.1. Materials
Phospholipid was purchased from Avanti (Shanghai, China); Cholesterol and glutaric anhydride were purchased from Aladdin (Shanghai, China); Sodium cholate was purchased from Tokyo Chemical Industry (Tokyo, Japan); Human apoA-I standard was purchased from Merck (Branchburg, NJ); FIV precipitate was kindly provided from Tonglu Biotechnology Co. Ltd. (Hangzhou, China); LCAT colorimetric assay kit was purchased from GenMed Scientifics Inc. (Arlington, MA); Goat anti-human apoA-I antibody and peroxidase-conjugated donkey anti-goat IgG were purchased from Abcam (Cambridge, UK). Mouse brain microvascular endothelial cells (bEND.3) and rat glioma cells (C6) were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The rest were commercially qualified reagents.
2.2. Extraction and identification of apolipoprotein A-I
2.2.1. Extraction of apoA-I
ApoA-I was mainly derived from FIV precipitation generated as the wastes produced in the industrial process of albumin. In this project, isothermal precipitation combined with organic solvent precipitation method was used to separate and extract apoA-I (Peitsch et al., Citation1989).Twenty grams FIV precipitation was dispersed in 200 mL phosphate-buffered saline (PBS, 0.02 M, pH 5.5). After adding absolute ethanol (final concentration 20%), the suspension was centrifuged for 25 min at 15,000 rpm and 4 °C. The deposit was collected and redispersed in 200 mL PBS (0.05 M, pH 6.6), then 20 mL anhydrous ethanol was added, and the second centrifugation procedure was carried out under the same conditions to obtain the crude HDL. The resulted supernatant was filtered through 0.8 µm filters. The filtrate was adjusted to pH 5.5 with PBS (1.65 M, pH 3.6) containing 18% ethanol, and the precipitate was centrifugally separated under the same conditions to obtain the expected HDL. HDL was fully dissolved in 1 mL Tris–HCl–urea solution and was dripped into 50 mL anhydrous ethanol with the subsequent delipidation for 12 h. The apoA-I precipitation collected after centrifugation was dissolved and dialyzed overnight in PBS (0.05 M, pH 6.6) to remove impurities. It was stored at −20 °C for subsequent experiment after lyophilization.
2.2.2. Identification of apoA-I
The apos and delipidated FIV precipitate were both identified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (Holmquist and Carlson, Citation1977). The gels were photographed with gel image system after completion of electrophoresis (GelDoc 2000, Bio-Rad, Hercules, CA). The molecular weight (MW) of extracted protein was estimated through linear regression between migrated distance and logarithm of MW of standard marker protein. ApoA-I bounded in the extracted apos was also confirmed by Western blotting. The sample separated using SDS-PAGE was electrophoretically transferred to membranes (100 V, 4 °C, 90 min). ApoA-I was detected with a polyclonal goat anti-human apoA-I antibody, accompanied by a horseradish peroxidase-conjugated donkey anti-goat IgG. Detection was performed using the enhanced chemiluminescence (ECL) method.
2.3. Synthesis and identification of MCG
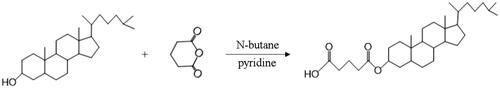
Glutaric anhydride was covalently attached to 3′-hydroxy of cholesterol to obtain the modified cholesterol according to synthetic route in . N-butane was chosen as reaction solvent to dissolve cholesterol and glutaric anhydride (the mass ratio was 3.39:1) in the round flask. After stirring for 20 min, 2 mL pyridine was added as catalyzer and the mixture was refluxed for 15 h. Then, cooling crystallization and suction filtration were carried out to obtain the crude product. After recrystallization and lyophilization, expected compound was collected as a white powder, followed by FI-IR and 1H NMR measurements to confirm the structure of the target product.
2.4. Synthesis and identification of MCG modified apoA-I (mA)
2.4.1. Synthesis
Carboxyl group of MCG was activated by EDC and NHS in dichloromethane at room temperature overnight to obtain MCG-NHS active ester (MCG-NHSE, the molar ratio was 1:2:2). MCG-NHSE was dissolved in 0.5 mL DMF and added dropwise to apoA-I solvent with agitation (the molar ratio was 150:1), followed by stirring overnight at 4 °C. The solvent was dialyzed against distilled water for 48 h and centrifuged at 12,000 rpm and 4 °C for 10 min. The expected mA was obtained after lyophilization.
2.4.2. Determination of the degree of substitution (DS) of mA
TNBS method was utilized to detect the DS of apoA-I (Laccotripe et al., Citation1997). ApoA-I and mA stock solution were diluted with PBS to obtain series samples with the concentrations in the range from 0.2 to 1.0 mg/mL. Each sample was mixed with 0.2 mL NaHCO3 solution (4% w/v, pH 8.5) and 0.2 mL TNBS solution (0.1%, w/v) and incubated at 40 °C for 2 h in the dark, followed by adding 0.2 mL 10% SDS and 0.1 mL 1 M HCl. The absorbance of samples at 420 nm was measured with a multi-mode microplate reader (ELx808, BioTek, Winooski, VT). The linear regression was conducted between the protein absorbance as Y-axis and the concentration as X-axis to obtain the standard curve and the regression equation. The DS was calculated by the slope of the equation according to the following formula:
(1)
(1)
where DS is the substitution degree of mA; KmA and KapoA-I are the slope rates of mA and apoA-I, respectively.
2.4.3. Average MW measurement of mA
The modified protein was subjected to SDS-PAGE electrophoresis to evaluate the MW by correlating the electrophoretic band position with marker. The DS of protein can also be deduced from the following formula:
(2)
(2)
where DS is substitution degree of mA; MWmA, MWapoA-I, and MWMCG are the relative molecular mass of mA, apoA-I, and MCG, respectively.
2.4.4. Surface tension estimation of mA
The surface tension of apoA-I and mA was also measured with DCAT 2.1 tension meter (Dataphysics, Filderstadt, Germany) (Bhuiyan et al., Citation2015). Briefly, stock solutions of apoA-I and mA were diluted with PBS to obtain a series of samples in the concentrations of 0.02–0.5 mg/mL prior to the measurements, and each sample was assayed in triplicate.
2.5. Preparation and characterization of d-rHDL
2.5.1. Preparation of d-rHDL
Lipid core of d-rHDL was prepared by thin-film dispersion method. Briefly, phospholipid and cholesterol with or without coumarin 6, a model drug with lipophilicity and fluorescent probe (the mass ratio was 30:5:1) were dissolved in methanol/dichloromethane (1:3, v/v) mixture and the solution was dried under a high vacuum at 50 °C using a rotary evaporator (RE2000B, Yarong Biochemical Instrument Factory, Shanghai, China). The lipid film was then rehydrated in 15 mL of 0.01 M PBS buffer (pH 7.4) containing 30 mg sodium cholate and the turbid emulsion was subsequently probe sonicated using a SCIENTZ-IID sonifier (SCIENTZ, Ningbo, China) at 250 W output for 3 min. After filtration through a millipore filter (0.22 μm), the lipid core suspension was obtained. Then, apoA-I (30 mg) suspended in 2 mL PBS buffer was added into the above emulsion and the mixture was further incubated at 4 °C for 6 h to obtain d-rHDL after dialysis (Oda et al., Citation2006; Hughes, Citation2009). Besides, mA constituted d-rHDL (m-d-rHDL) was prepared with the same method except for the alternative of mA for cholesterol and apoA-I.
2.5.2. Morphology, size, and zeta potential
Morphology of m-d-rHDL before and after incubation with mA was observed under TEM (FEI Company, Hillsboro, OR). Briefly, the lipid core and m-d-rHDL were dropped onto a copper grid, then negatively stained with 1% uranyl acetate and naturally evaporated at room temperature. Size distribution and zeta potential were measured using NICOMP 380/ZLS Zeta potential/particle size analyzer (PSS.NICOMP Particle Systems, Santa Barbara, CA).
2.5.3. Determination of coumarin 6 loading capacity (LC) and encapsulation efficiency (EE)
LC and EE of coumarin 6 in d-rHDL and m-d-rHDL were quantified using HPLC analysis (Shimadzu LC-20A, Kyoto, Japan) equipped with a fluorescence detector set at Ex 465 nm/Em 502 nm. The separation was performed at 30 °C on a Synergi Hydro-RP C18 column (5 μm, 250 mm × 4.6 mm, Phenomenex, Torrance, CA) protected by a C18 Securityguard column (5 μm, 10 mm × 4.6 mm, Kromasil, Bohus, Sweden). The mobile phase was acetonitrile and water in the ratio of 95:5 (v/v) and delivered at a flow rate of 1.0 mL/min (Xu et al., Citation2010). Mini-column centrifugation method was applied to separate coumarin 6 loaded d-rHDL or m-d-rHDL from free coumarin 6 (Chu et al., Citation2014). In brief, d-rHDL or m-d-rHDL suspension was added dropwise to the center of Sephadex G50 mini-column, followed by the centrifugation for 5 min at 1000 rpm. d-rHDL or m-d-rHDL particles could be collected through the above centrifugal separation process while the free coumarin 6 was still remained in the Sephadex G50 mini-column. After demulsification with absolute ethanol, the amount of coumarin 6 entrapped in the d-rHDL or m-d-rHDl and the total amount of coumarin 6 in the suspension were determined by HPLC, respectively. EE and LC were calculated according to the following formulas:
(3)
(3)
where EE is the encapsulation efficiency; W and W0 are the amount of encapsulated coumarin 6 and the total amount of coumarin 6 in the suspension, respectively.
(4)
(4)
where WC is the amount of coumarin 6 loaded in the nanoparticles; WT is the total amount of nanoparticles and the loaded coumarin 6.
2.5.4. Plasma colloidal stability assay
To demonstrate the plasma colloidal stability of d-rHDL and m-d-rHDL under physiological condition, variations in particle size and PDI value were determined in the presence of PBS 7.4 containing 10% FBS. In brief, d-rHDL and m-d-rHDL were suspended at a concentration of 10% FBS solution in PBS 7.4 at 37 °C, 100 rpm. At predetermined time points (0, 1, 2, 4, 8, 12, and 24 h), the particle sizes and PDI values of d-rHDL and m-d-rHDL were measured with NICOMP 380/ZLS Zeta potential/particle size analyzer.
2.6. Allosteric behavior of d-rHDL
2.6.1. Extraction of LCAT
LCAT was extracted with reference to the method provided by Holmquist (Laccotripe et al., Citation1997) as follows: the human plasma was mixed with 3 μg/mL glacial acetic acid at 0 °C with equal volume, then the diluted plasma was centrifuged at 5000 rpm and 0 °C for 15 min. The supernatant was added to the Q-agarose gel column pretreated with N-methyl piperazine buffer solution (pH 5.0, 20 mM), and the column was washed with the buffer followed by real-time monitoring at 280 nm with nucleic acid protein analyzer (QiTe, Shanghai, China) until no protein could be detected in the effluent. Eventually, the adsorbed LCAT was eluted at 2.2 mL/min with 0.5 mol/L NaCl at 4 °C. The collected eluent was dialyzed to remove NaCl and then condensed to its half original volume. The resulting solvent was centrifugally separated to obtain the expected LCAT.
2.6.2. Microscope observation
0.1 mL d-rHDL and m-d-rHDL were incubated with 0.1 mL LCAT for 2 h at 37 °C, respectively. Then, the samples were added on the copper grid and then negatively stained with 1% uranyl acetate. After drying at room temperature, TEM was employed to observe and compare the morphological differences between m-d-rHDL and d-rHDL after treatment with LCAT.
2.6.3. Release behavior investigation
Release behavior of coumarin 6 from m-d-rHDL and d-rHDL in vitro was performed to clarify the effect of structural modification on drug leakage. 0.5 mL m-d-rHDL and d-rHDL treated with 0.5 mL LCAT were respectively transferred into dialysis bag, then immersed in 200 mL pH 7.4 PBS buffer solution containing 10% FBS, and stirred gently at 37 °C and 100 rpm. The dialysis medium was withdrawn at fixed time, and fresh buffer was simultaneously supplemented. Samples collected at each time were filtered through 0.22 μm microporous membrane. Continuous filtrate was served for determination by HPLC. The release amount was calculated and the release curve was drawn. Release behavior of m-d-rHDL and d-rHDL without LCAT addition was also investigated in the same way.
2.7. Cytotoxicity and cell uptake test in vitro
2.7.1. Cell culture
Mouse brain microvascular endothelial cells (bEND.3) and rat glioma cells (C6) were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% inactivated fetal bovine serum (FBS) and 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C under humidified environment of 5% CO2 (Thermo Fisher, Waltham, MA).
2.7.2. Cytotoxicity assay in vitro
CCK-8 assay was conducted to estimate the cell toxicities of lipid core, d-rHDL and m-d-rHDL. bEND.3 were seeded in 96-well plates at a density of 1 × 104 cells/well and cultured for 24 h. One hundred microliters lipid core, d-rHDL, and m-d-rHDL were added to each well diluted with DMEM to final suspension concentrations of 10 μg/mL, 500 μg/mL, and 1000 μg/mL, respectively. Control group and blank group were considered and set up simultaneously. DMEM suspension was removed after incubating for 24 h, then 100 μL CCK-8 working solution was added to each well and returned to the incubator for 1 h. The UV absorbance was measured using a multi-mode microplate reader at 450 nm. Cytotoxicities of lipid core and m-d-rHDL on C6 cells were also evaluated as described above. Cell viability was calculated by the following equation:
(5)
(5)
where ODexperiment well, ODcontrol well, and ODblank well are the optical density of experiment well, control well, and blank well, respectively.
2.7.3. Cell uptake
bEND.3 and C6 cells were seeded into 24-well plates at a density of 2 × 104 cells/well and cultured for 24 h at 37 °C. In order to investigate whether or not there is any difference between d-rHDL and m-d-rHDL in cell uptake under the action of LCAT, coumarin 6-labeled lipid core, d-rHDL and m-d-rHDL were added to bEND.3 cells in the presence of LCAT (100 μg/mL) and incubated for 1 h, 2 h, and 4 h, respectively. After incubation, the cells were rinsed three times with PBS and fixed with 4% paraformaldehyde for 10 min. The phagocytized nanoparticles were visualized under a inverted fluorescence microscope (Olympus Corporation, Tokyo, Japan) and quantitatively measured with a multi-mode microplate reader (Ex: 466 nm, Em: 504 nm). In addition, the internalization of coumarin 6 loaded lipid core and m-d-rHDL by C6 cells was further assayed in the absence of LCAT for 1 h, 2 h, and 4 h.
2.8. Statistical analysis
Statistically significant differences were utilized using two-tailed Student’s t-test or one-way ANOVA (SPSS, version 16.0, SPSS Inc., Chicago, IL). Statistical significance was noted as follows: *p<.05; **p<.01; ***p<.001.
3. Results and discussion
3.1. Identification of apoA-I
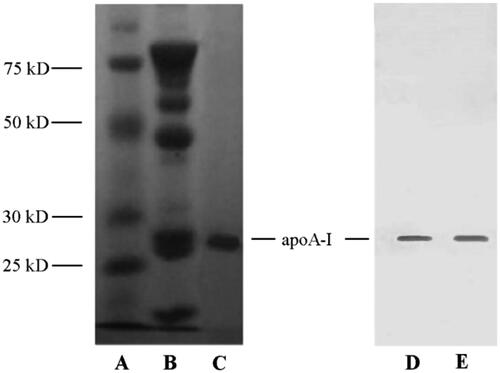
ApoA-I was extracted from human blood albumin waste FIV precipitation referring to the cold-ethanol isoelectric precipitation procedure (Song et al., Citation2014) and identified through SDS-PAGE (, left) and Western blot assay (, right). In contrast to FIV precipitation (), SDS-PAGE pattern of extracted products exhibited a primary band around 28.7 kDa without any other irrelevant protein in , which was consistent with the MW of apoA-I. Positive Western blot presentation provided further evidence that apoA-I was identified to be abounded in extracted products () and could be regarded as primary protein from human blood albumin waste FIV precipitation.
3.2. Identification of MCG
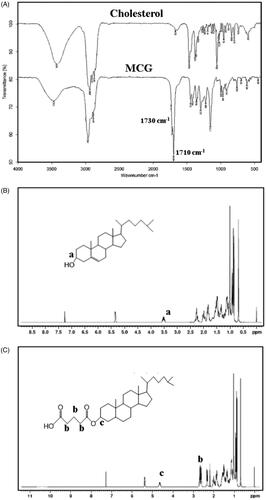
The molecular structure of MCG was verified through FI-IR and 1H NMR tests. As illustrated in , FT-IR spectra of MCG exhibited a characteristic peak around 1710 cm−1 corresponding to carboxy carbonyl group, followed by a peak at 1730 cm−1, which may be ascribed to ester carbonyl group. In , chemical shift from 2.62 ppm to 2.70 ppm was observed corresponding to methylene hydrogen (–OCOCH2CH2CH2COOH) between ester carbonyl and carboxy carbonyl groups. Owing to the esterification of glutaric anhydride and cholesterol, chemical shift of methyl group moved from 3.53 ppm in cholesterol 1H NMR () to 4.65 ppm in MCG 1H NMR (). Based on the above structure validation results, MCG was ascertained to be formed and the synthetic procedure was successful and feasible. The yield of MCG was calculated as 72.65%, and the purity was 93.32%.
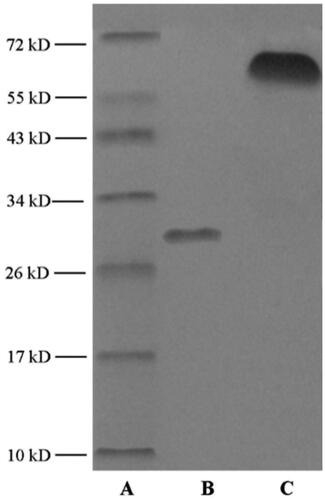
3.3. Characterization of mA
As presented in , different migration was observed in SDS-PAGE of mA compared with apoA-I. According to marker image, MW of mA was estimated to about 67 kDa. The remarkable increment indicated that MCG was covalently conjugated to apoA-I. The DS of mA was estimated to be about 65% according to EquationEquation (1)(1)
(1) . Similar DS was speculated through TNBS method as well referring to EquationEquation (2)
(2)
(2) .
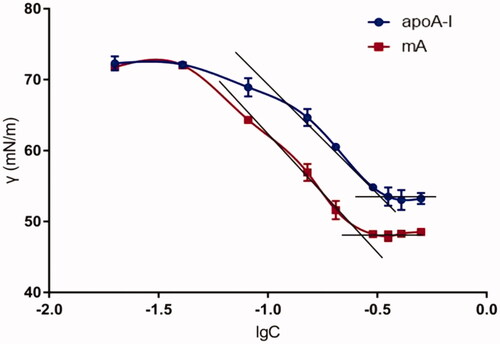
presents the diagram of surface tension (γ) against concentration (C) for apoA-I and mA. The minimum surface tension (γmin) obtained by inflection point of apoA-I was about 53.27 mN/m, and that of mA was about 48.78 mN/m (). The decrease of surface tension showed that amphiphilicity of apoA-I was improved through combining with MCG, which could facilitate the insertion of mA into the lipid core.
3.4. Characterization of m-d-rHDL
The structure transformation under TEM observation and the changes in particle size and zeta potential are clarified in and . Comparing to , m-d-rHDL cores () appeared as hollow vesicles surrounding with fingerprints-like structure and transformed into compactly arranged discoidal HDL stacks after incubation with mA (). The disappearance of internal water-cavity was due to the progressive deformation under the compression pressure exerted by mA to lipid core. For the same reason, a significant reduction in particle size from (89.23 ± 1.21) nm to (25.62 ± 2.34) nm and a slight decrease in EE were simultaneously measured (). All of the structural variations indicated the reappearance of a typical morphology of nascent HDL and the crucial and similar role of mA by replacing apoA-I to the transformation. After mA coupling, m-d-rHDL got a significant increment (about 49%) over the lipid cores in negative surface charge, which could not only be beneficial to its stability and dispersibility, but also again proved the anchor of mA on the surface of the lipid cores.
Figure 6. Transmission electron microscope of lipid core (A) and m-d-rHDL (B). The bar of A was 100 nm. The bar of B was 20 nm.
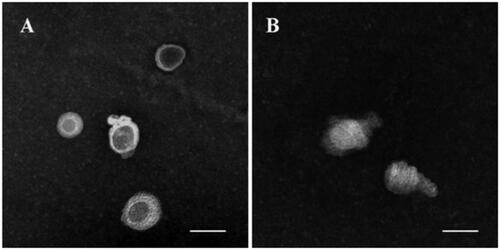
Table 1. Size, zeta potential, EE and LC of lipid core and m-d-rHDL (n = 3).
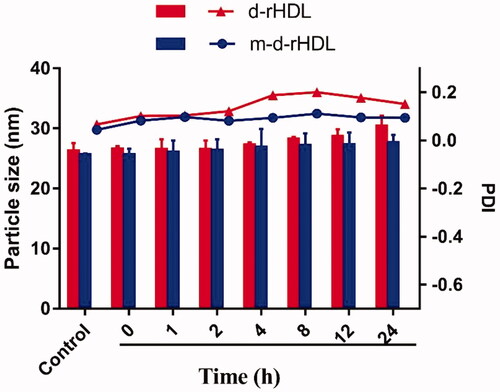
The colloidal stability of m-d-rHDL and d-rHDL under physiological condition was investigated through particle size assessment. As illustrated in , the particle size of m-d-rHDL and PDI value had no remarkable change in 24 h when incubated with 10% FBS solution, indicating a favorable colloidal stability and resistance to serum protein adsorption after apoA-I modification. However, the particle size of d-rHDL presented an approximate 15% increase for 24 h. Besides, PDI values fluctuation of d-rHDL (increase at first and then decrease) also proved the probability of a coexistence of multi-morphologies (spherical nanoparticles with disk stacks) after suspending in 10% FBS, which could be speculated that d-rHDL might have experienced an allosteric reaction when exposed to LCAT in FBS environment.
3.5. Allosteric behavior of d-rHDL
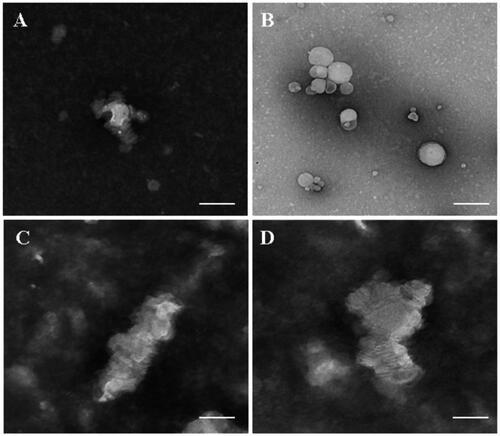
According to the mensuration with LCAT colorimetric assay kit, LCAT activity was 8.105 nmol/min/mL and was in good agreement with the certified values (Holmquist, Citation2002). The structure remodeling was evaluated by TEM observation and drug release behavior. Shown from , d-rHDL assembling with apoA-I and m-d-rHDL assembling with mA all appeared as disk-like structures in absence of LCAT. Nevertheless, most of d-rHDL transformed into round shape without central water cavity after mixing with LCAT for 2 h. The structure rearrangement of d-rHDL was consistent with previous studies (Kostner et al., Citation1987; Asztalos et al., Citation2007) and might be attributed to the conversion from free cholesterol to CE, the resultant compression and occupation of CE to the internal hydrophilic cavity, and the final formation of spherical rHDL.
Figure 8. Microphotographs of different preparations using transmission electron microscope. (A) d-rHDL without LCAT; (B) d-rHDL incubation with LCAT; (C) m-d-rHDL without LCAT; (D) m-d-rHDL incubation with LCAT. The bar of A was 50 nm. The bar of B was 100 nm. The bar of C and D was 20 nm.
Release behavior of coumarin 6 was further performed to assess and compare the allosteric behavior of d-rHDL and m-d-rHDL. LC of coumarin 6 in d-rHDL and m-d-rHDL was measured (0.0734 ± 0.005)% and (0.0663 ± 0.003)%, respectively (), and it was sufficient for the following investigation and determination in release behavior and cell uptake assays on account of its efficient quantum yield. illustrates the cumulative release profiles of coumarin 6 from d-rHDL and m-d-rHDL with or without LCAT at pH 7.4 PBS. The release percentages of coumarin 6 from d-rHDL treated with LCAT reached to 19.82%, 43.32%, and 66.23% at 4 h, 8 h, and 24 h, respectively, significantly faster than d-rHDL without LCAT at the corresponding time, indicating potential drug leakage in blood circulation and decreased targeting efficiency transferring to brain.
Figure 9. In vitro release profiles of coumarin 6 from d-rHDL and m-d-rHDL with or without LCAT under PBS 7.4 containing 10% FBS at 37 °C (n = 3).
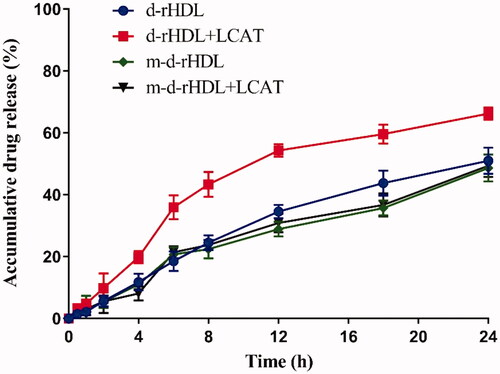
On the contrary, the original disk-shaped structure of m-d-rHDL was well remained even under the same LCAT-treated condition (). In terms of release behavior, m-d-rHDL appeared satisfactory sustained release characteristic totally different from d-rHDL with LCAT and had no significant changes whether in the presence or absence of LCAT (). It can be inferred from this obvious difference that cholesterol esterified with glutaric anhydride could effectively protect m-d-rHDL from LCAT catalysis and refrain from drug leakage prior to migrating into brain.
3.6. Cytotoxicity evaluation
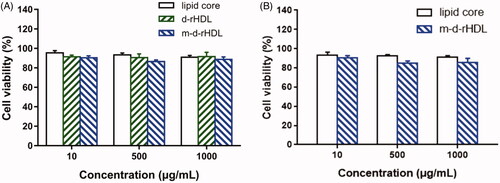
The in vitro cell viabilities of lipid core, d-rHDL, and m-d-rHDL are illustrated in . More than 90% of cells were viable at all concentrations ranging from 10 to 1000 μg/mL. Hence, lipid core, d-rHDL, and m-d-rHDL exhibited negligible cytotoxicity and highly favorable biocompatibility toward bEND.3 and C6 cells within all the tested concentrations.
3.7. Cell uptake study
In our study, bEND.3 monolayer model was constructed in vitro to evaluate the potential ability to transport through BBB (Yang et al., Citation2014). The internalization of coumarin 6 loaded lipid core, d-rHDL and m-d-rHDL by bEND.3 cells was visualized and quantified. All of the lipid core, d-rHDL and m-d-rHDL exhibited time-dependent internalization within 4 h of incubation ( and ). Observed under fluorescence microscope shown in , coumarin 6 labeled d-rHDL () and m-d-rHDL () by bEND.3 cells had much stronger fluorescent intensity compared with lipid core without apoA-I and mA involved () at all the corresponding times, about 1.32-fold and 2.46-fold more than that of lipid core at 4 h, respectively (). This may be due to the brain-targeting property of apoA-I anchored on the surface of rHDL associated with SR-BI specifically expressed on brain microvascular endothelial cells (Gong et al., Citation2019). It suggested that the uptake of nanoparticles in bEND.3 cells could be improved by the binding with apoA-I. As shown in , when the cells were pre-incubated with LCAT, a significant inhibition effect on the uptake of d-rHDL was observed. When compared to m-d-rHDL, the relative uptake intensity decreased by about 16% at 1 h, 30% at 2 h and 47% at 4 h, separately. Together with the above release behavior difference, this result reconfirmed the negative impact of rHDL remodeling on cellular internalization and the positive effectiveness on improving cellular uptake by protecting m-d-rHDL from LCAT catalysis and avoiding from drug leakage. Based on the aforementioned results, m-d-rHDL possessed more efficient targeting activity toward bEND.3 cells compared with d-rHDL and could be regarded as a potential vehicle to traverse the BBB mediated by modified apoA-I.
Figure 11. Fluorescence images of bEND.3 cells after incubation with lipid core (A–C), d-rHDL (D–F), and m-d-rHDL (G–I) at 37 °C for 1 h, 2 h, and 4 h, respectively.
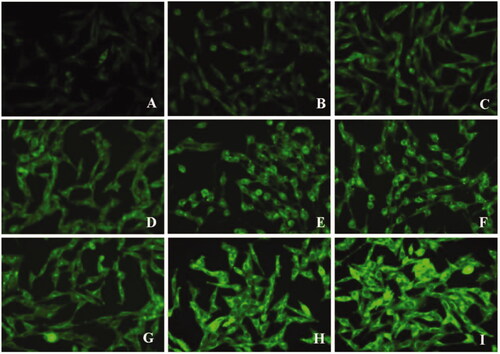
Figure 12. Fluorescence images of C6 cells after incubation with lipid core (A–C) and m-d-rHDL (D–F) at 37 °C for 1 h, 2 h, and 4 h, respectively.
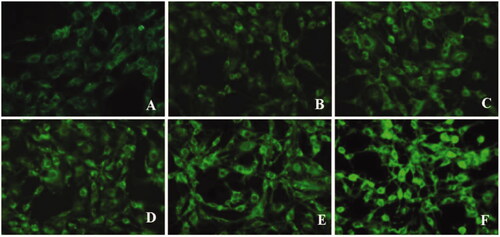
Figure 13. Uptake of lipid core, d-rHDL and m-d-rHDL in bEND.3 cells (A) and C6 cells (B) observed qualitatively by a multi-mode microplate reader (n = 5), *p<.05, **p<.01.
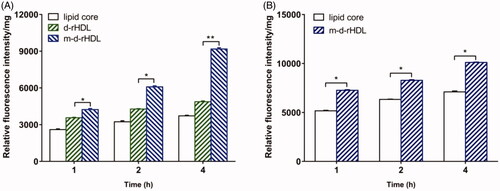
Moreover, C6 cells were utilized to simulate glioma model in vitro (Eavarone et al., Citation2000). Similar to the result in qualitative analysis (), the cellular uptake of m-d-rHDL by C6 cells was prominently enhanced quantitatively as it was compared with the lipid core at every measured timepoint (). The favorable penetration behavior of m-d-rHDL confirmed the important role of apoA-I in promoting C6 cells uptake of more m-d-rHDL by specific receptors expressed on the surface of glioma cells.
4. Conclusions
Reconstituted HDL offers great promise as an effective targeting nanocarrier to facilitate the delivery of drugs toward glioma cells, mainly via the mechanism of apoA-I mediation. However, in view of the drug leakage resulting from the remodeling of d-rHDL and the consequent reduced targeting efficiency under the action of LCAT, a novel HDL-mimic nanoparticle was developed, which was composed of a liposomal core and a MCG modified apoA-I shell. The obtained nanocarrier presented disk stacks with similar properties such as shape, size range, and composition resembling native nascent HDL. Both the colloidal stability and allosteric behavior including microscope observation and in vitro release behavior proved that the remolding tendency of d-rHDL was successfully suppressed through the structural modification of cholesterol. The uptake of m-d-rHDL in LCAT-pretreated bEND.3 cells significantly increased with that of d-rHDL, which would be another favorable evidence for its capability of improving targeting efficiency by protecting m-d-rHDL from LCAT catalysis and avoiding from drug leakage. Besides, apoA-I anchored on the surface of d-rHDL and m-d-rHDL played an important role in SR-BI meditated endocytosis process of m-d-rHDL into bEND.3 cells and C6 cells. Therefore, this novel HDL-mimic nanoparticle assembled with mA was engineered to effectively overcome the remolding drawback of disk-rHDL and provide highly desirable physicochemical characteristics as a brain-targeted delivery nanoplatform for improving therapeutic effect.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Asztalos BF, Schaefer EJ, Horvath KV, et al. (2007). Role of LCAT in HDL remodeling: investigation of LCAT deficiency states. J Lipid Res 48:592–9.
- Bhuiyan MHU, Saidur R, Mostafizur RM, et al. (2015). Experimental investigation on surface tension of metal oxide–water nanofluids. Int Commun Heat Mass Transfer 65:82–8.
- Brandner S, Jaunmuktane Z. (2018). Neurological update: gliomas and other primary brain tumours in adults. J Neurol 265:717–27.
- Carlucci A, Cigliano L, Maresca B. (2012). LCAT cholesterol esterification is associated with the increase of ApoE/ApoA-I ratio during atherosclerosis progression in rabbit. J Physiol Biochem 68:541–53.
- Chen R, Cohen AL, Colman H. (2016). Targeted therapeutics in patients with high-grade gliomas: past, present, and future. Curr Treat Options Oncol 17:42.
- Chu Y, Li D, Luo YF, et al. (2014). Preparation and in vitro evaluation of glycyrrhetinic acid-modified curcumin-loaded nanostructured lipid carriers. Molecules 19:2445–57.
- Cooke AL, Morris J, Melchior JT, et al. (2018). A thumbwheel mechanism for APOA1 activation of LCAT activity in HDL. J. Lipid Res 293:5462–3.
- Cui L, Wang Y, Liang M, et al. (2018). Dual-modified natural high density lipoprotein particles for systemic glioma-targeting drug delivery. Drug Deliv 25:1865–76.
- Eavarone DA, Yu X, Bellamkonda RV. (2000). Targeted drug delivery to C6 glioma by transferrin-coupled liposomes. J Biomed Mater Res 51:10–4.
- Florence AT, Halbert GW. (1991). Lipoproteins and microemulsions as carriers of therapeutic and chemical agents. Targeted Diagn Ther 5:141–74.
- Gong M, Zhang Q, Zhao Q, et al. (2019). Development of synthetic high-density lipoprotein-based ApoA-I mimetic peptide-loaded docetaxel as a drug delivery nanocarrier for breast cancer chemotherapy. Drug Deliv 26:708–16.
- Gordts SC, Singh N, Muthuramu I, Geest BD. (2013). Pleiotropic effects of HDL: towards new therapeutic areas for HDL-targeted interventions. Curr Mol Med 14:6744–53.
- Holmquist L. (2002). Selective extraction of lecithin:cholesterol acyltransferase (EC 2.3.1.43) from human plasma. J Biochem Biophys Methods 52:64–8.
- Holmquist L, Carlson K. (1977). Selective extraction of human serum very low density apolipoproteins with organic solvents. Biochim Biophys Acta 493:400–9.
- Hong SC, Zhao SP, Wu ZH. (2007). Effect of probucol on HDL metabolism and class B type I scavenger receptor (SR-BI) expression in the liver of hypercholesterolemic rabbits. Int J Cardiol 115:29–35.
- Hughes DP. (2009). Expert opinion on drug delivery: strategies for the targeted delivery of therapeutics for osteosarcoma. Expert Opin Drug Deliv 6:1311–21.
- Kostner GM, Knipping G, Groener JEM, et al. (1987). The role of LCAT and cholesteryl ester transfer proteins for the HDL and LDL structure and metabolism. Adv Exp Med Biol 210:79–86.
- Kratzer I, Wernig K, Panzenboeck U, et al. (2007). Apolipoprotein A-I coating of protamine-oligonucleotide nanoparticles increases particle uptake and transcytosis in an in vitro model of the blood–brain barrier. J Control Release 117:301–11.
- Laccotripe M, Makrides SC, Jonas A, Zannis VI. (1997). The carboxyl-terminal hydrophobic residues of apolipoprotein A-I affect its rate of phospholipid binding and its association with high density lipoprotein. J Biol Chem 272:17511–22.
- Lavrador JP, Oliveira E, Pimentel J, Livraghi S. (2017). Adult pilocytic astrocytoma of conus medullaris: clinical considerations and review of the literature. CNS Oncol 6:107–10.
- Lin L, Cai J, Jiang C. (2017). Recent advances in targeted therapy for glioma. Curr Med Chem 24:1365–81.
- Lu H, Zhang H, Zhang D, et al. (2015). A biocompatible reconstituted high-density lipoprotein nano-system as a probe for lung cancer detection. Med Sci Monit 21:2726–33.
- Lund-Katz S, Liu L, Thuahnai ST, Phillips MC. (2003). High density lipoprotein structure. Front Biosci 8:d1044–d54.
- Marsche G. (2015). It’s time to reassess the high-density lipoprotein (HDL) hypothesis: CSL112, a novel promising reconstituted HDL formulation. J Am Heart Assoc 4:68–76.
- Nicholls SJ, Puri R, Nissen SE. (2018). High-density lipoprotein-targeted therapies-not dead yet-reply. JAMA Cardiol 3:1255–6.
- Nobecourt E, Davies MJ, Brown BE, et al. (2007). The impact of glycation on apolipoprotein A-I structure and its ability to activate lecithin:cholesterol acyltransferase. Diabetologia 50:643–53.
- Oda MN, Hargreaves PL, Beckstead JA, et al. (2006). Reconstituted high density lipoprotein enriched with the polyene antibiotic amphotericin B. J Lipid Res 47:260–7.
- Park KH, Yun CO, Kwon OJ, et al. (2010). Enhanced delivery of adenovirus, using proteoliposomes containing wildtype or V156K apolipoprotein A-I and dimyristoylphosphatidylcholine. Hum Gene Ther 21:579–87.
- Pedersbaek D, Kraemer MK, Kempen PJ, et al. (2019). The composition of reconstituted high-density lipoproteins (rHDL) dictates the degree of rHDL cargo- and size-remodeling via direct interactions with endogenous lipoproteins. Bioconjug Chem 30:2634–46.
- Peitsch MC, Kress A, Lerch PG, et al. (1989). A purification method for apolipoprotein A-I and A-II. Anal Biochem 178:301–5.
- Petri B, Bootz A, Khalansky A, et al. (2007). Chemotherapy of brain tumour using doxorubicin bound to surfactant-coated poly(butyl cyanoacrylate) nanoparticles: revisiting the role of surfactants. J Control Release 117:51–8.
- Qian W, Qian M, Wang Y, et al. (2018). Combination glioma therapy mediated by a dual-targeted delivery system constructed using OMCN-PEG-Pep22/DOX. Small 14:123–32.
- Rajora MA, Zheng G. (2016). Targeting SR-BI for cancer diagnostics, imaging and therapy. Front Pharmacol 7:326.
- Song Q, Huang M, Yao L, et al. (2014). Lipoprotein-based nanoparticles rescue the memory loss of mice with Alzheimer’s disease by accelerating the clearance of amyloid-beta. ACS Nano 8:2345–59.
- Tosheska Trajkovska K, Topuzovska S. (2017). High-density lipoprotein metabolism and reverse cholesterol transport: strategies for raising HDL cholesterol. Anatol J Cardiol 18:149–54.
- Tsompanidi EM, Brinkmeier MS, Fotiadou EH, et al. (2010). HDL biogenesis and functions: role of HDL quality and quantity in atherosclerosis. Atherosclerosis 208:3–9.
- Vaisar T, Pennathur S, Green PS, et al. (2007). Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest 117:746–56.
- Wang J, Jia J, Liu J, et al. (2013). Tumor targeting effects of a novel modified paclitaxel-loaded discoidal mimic high density lipoproteins. Drug Deliv 20:356–63.
- Xu Y, Jin X, Ping Q, et al. (2010). A novel lipoprotein-mimic nanocarrier composed of the modified protein and lipid for tumor cell targeting delivery. J Control Release 146:299–308.
- Yang ZZ, Li JQ, Wang ZZ, et al. (2014). Tumor-targeting dual peptides-modified cationic liposomes for delivery of siRNA and docetaxel to gliomas. Biomaterials 35:5226–39.
- Yang F, Zou Y, Gong Q, et al. (2020). From astrocytoma to glioblastoma: a clonal evolution study. FEBS Open Bio 56:23–32.
- Zhang Y. (2004). Intravenous RNA interference gene therapy targeting the human epidermal growth factor receptor prolongs survival in intracranial brain cancer. Clin Cancer Res 10:3667–77.
- Zhu C, Kros JM, Cheng C, Mustafa D. (2017). The contribution of tumor-associated macrophages in glioma neo-angiogenesis and implications for anti-angiogenic strategies. Neuro Oncol 19:1435–46.