Abstract
In this study, a rapid and highly specific TaqMan-based duplex real-time polymerase chain reaction method, based on the simultaneous amplification of fragments of the mitochondrial ND2 and ND5 genes, was developed and optimized for the identification and quantification of pork and donkey meats in raw and cooked binary donkey/beef and pork/beef mixtures. Accumulation of polymerase chain reaction products was monitored by measuring the fluorescent signals from FAM and HEX labeled probes specific for pork and donkey, respectively. As a consequence, target meat species could be detected at a level of 0.001% in raw and oven-cooked meat mixtures, and at a level of 0.01% in autoclaved meat mixtures. The result in this study indicated that the TaqMan-based duplex polymerase chain reaction assay could be successfully used in the identification and quantification of donkey and pork in adulteration studies with a high degree of specificity and sensitivity.
INTRODUCTION
Fraudulent substitution or undeclared adulteration of meat species in comminuted and highly processed meat products is a widespread problem in retail markets.[ Citation 1 ] Undesired practices include replacement of high commercial value meat species with cheaper or undesired meats, the adulteration in non-meat (vegetable proteins) products, and utilization of meat in lower quantities than shown on the product label. Several analytical techniques have been developed for the qualitative and quantitative determination of such replacements in mixed meat mixtures. These methods include different protein-based techniques and related methods, such as electrophoretic techniques,[ Citation 2 ] enzyme-linked immuno-sorbent assays (ELISA),[ Citation 3 ,Citation 4 ] and isoelectric focusing (IEF).[ Citation 5 ] Recently, DNA-based techniques could offer the potential to identify a particular animal species because of unique stability and specificity of DNA.[ Citation 6 ,Citation 7 ] Polymerase chain reaction (PCR) is a fast, sensitive, and highly specific technique and it could be used as an alternative to protein-based methods.[ Citation 8 ] However, when a conventional PCR technique is used, it only allows for a qualitative identification of the species, not quantitative.
Nowadays, the real-time PCR technique is a very common tool for species identification and quantification because of its high sensitivity and specificity, larger dynamic detection range, and less carry-over contamination.[ Citation 9 − Citation 12 ] In this respect, the real-time PCR method now appears to be the most effective and robust technique for the identification and quantification of trace quantities of different animal species, even in complex foods.[ Citation 13 − Citation 15 ] Currently, two approaches have been employed to obtain a flourescent signal from the PCR products. The first approach involves the use of intercalating fluorescent dyes like SYBR Green.[ Citation 16 ,Citation 17 ] In the SYBR Green method, these dyes bind to all double stranded DNA, also to any non-specific PCR products and to the primer-dimer complex because of the specifity depending on only two PCR primers. The second method is considered to be a more accurate and reliable method in which fluorescent reporter probes like the TaqMan fluorogenic probe system are used.[ Citation 18 − Citation 24 ] Unlike the SYBR Green method, an additional oligonucleotide is used in the TaqMan probe system. This oligonucleotide is a probe with both a reporter fluorescent dye and a quencher dye attached. In this system, the probe anneals to its complementary target sequence during the annealing step. During the extension stage, the probe is cleaved by the 5′ nuclease activity of the Taq DNA-polymerase and then the fluorescence signal increases.[ Citation 25 ]
The TaqMan-based real-time PCR technique has been used so far in some studies for the identification and quantification of animal-derived materials in meat mixtures. However, the determination of several meat species using simplex PCR approaches is laborious, expensive, and time consuming. In addition, higher rates of error may be unavoidable by applying single PCR systems, which also require many pipetting steps. In contrast, duplex systems are carried out in only one PCR reaction, therefore, saving time and cost. Furthermore, the results obtained in multiplex systems are not necessarily compiled; they can be collected at once.[ Citation 26 ] In Turkey, donkey and pork meats are generally used for fraudulent substitutions in meat products. Therefore, in this study, a TaqMan-based duplex real-time PCR approach was developed for simultaneous identification and quantification of donkey and pork adulteration in raw and heat-processed meats.
MATERIALS AND METHODS
Sample Preparation
In this research, donkey (Equus asinus), porcine (Sus scrofa), and bovine (Bos tauros) muscle tissues were used in the preparation of the binary meat mixtures. Donkey, pork, and beef samples were obtained from Research and Application center of the Veterinary Faculty in Erciyes University, Bonus External Trade Inc. Co., and a local butchery in Kayseri, respectively. The meat samples were transferred into the laboratory under cold chain and kept at −20°C until analyzed. To prepare the binary meat mixtures, donkey meat and pork were separately incorporated into beef at levels ranging from 0.001 to 10% (corresponding range of 0.001–10 mg/kg). In order to achieve homogeneity and eliminate errors during blending, beef was added to each binary meat mixture at a level of approximately 50% of the mixture and blended for 2 min to obtain completely homogeneous meat mixtures each time. Then, 20 g of each mixture was shaped into patty samples of 4 cm in diameter and 0.8 cm in thickness by using a metal shaper. After the preparation of the raw samples, they were either autoclaved (Hirayama/HV 50 L/Japan) at 120°C for 30 min (sterilization temperature conditions for canned meats) or cooked on aluminum plates in a drying oven (Memmert/INE 200-800, Germany) at 200°C for 30 min (baking temperature for canned meat meals). Then, DNA isolation was made from these raw and cooked binary meat mixture samples.
DNA Extraction
In order to obtain representative specimens, approximately 10–15 g of raw and cooked meat patty samples and pure meats from each species investigated were sampled and then powdered after holding at −80°C overnight. The DNA was extracted from 25 mg of powdered samples using a commercial kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. The concentration and purity of the extracted DNAs were determined by the ratio of the absorbance at 260 and 280 nm.[ Citation 27 ]
Duplex Real-Time PCR Assay
Duplex real-time PCR was performed as recently published, targeting the donkey specific region of the mtND2 gene and the pork specific region of the mtND5 gene.[ Citation 12 ] Primers and probes were purchased from Metabion (Martinsried, Germany). The TaqMan PCR reaction was performed in a final volume of 25 μl using 12.5 μl of Quantitect Probe PCR mix (Qiagen, Hilden, Germany,) 100 ng template DNA, 0.8 μM of sense and antisense primer, and 0.25 μM of the TaqMan probe for each species. The sense, antisense primers and probes used for donkey specific real-time PCR were KF: 5′-TGCTAGCCTCATTATCAGTAT-3′, KR: 5′-GTGATGAGGATACGTGCT-3′ and 5′-HEX-TCTACCAATCATATCATCAATCCTCAAC-TAMRA-3′, respectively. For pork specific real-time PCR, the sense, antisense primer, and probes used were DF: 5′-TACACTACCCTTATCATAACAG-3′, DR: 5′-ATACTGGGATTATTGCTAATAG-3′, and 5′-FAM-AATGTCCGGAACCATACTAGTAATAATC-TAMRA-3′. The amplifications were performed on a thermal cycler Line Gene II PCR device (Bioer Technology Co., Hangzhou, China) and a thermal cycling protocol, consisting of 95°C for 15 min followed by 40 cycles at 95°C for 15 s and at 60°C for 1 min. Reactions were replicated at least three times per experiment and experiments were replicated twice to verify the results.
Specificity of Duplex Real-Time PCR System
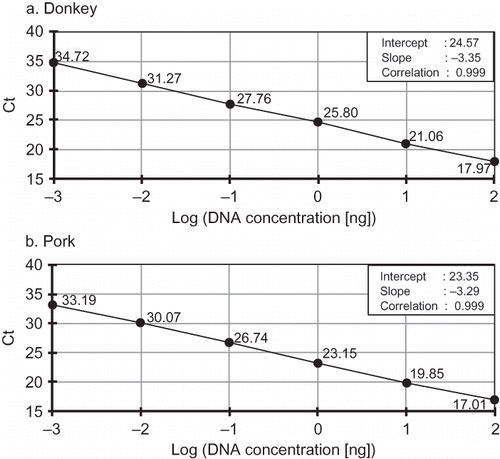
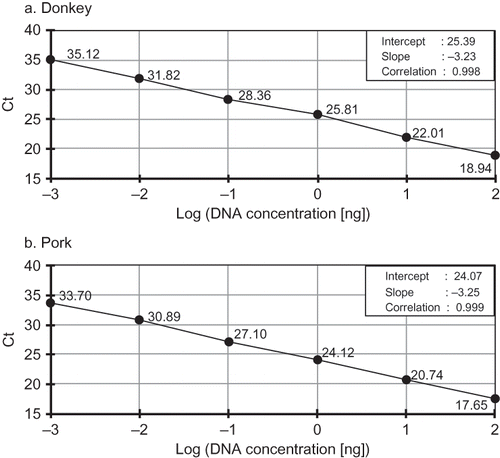
The specificity test of the duplex real-time PCR assay was carried out by amplification of 100 ng of donkey, pork, horse, bovine, ovine, turkey, and chicken genomic DNA. The specificity results obtained from duplex assay were compared with those obtained from the simplex assay. Each assay was tested against DNA from all seven species, i.e., against its target species and the five remaining species to confirm assay specificity. In the determination of the detection limit of the specific primers and probes, 1:10 serial reference dilutions of donkey and pork genomic DNAs were prepared. In a simplex PCR assay, 100, 10, 1, 0.1, 0.01, or 0.001 ng/μl DNA of either donkey or pork was separately added to each of the 25 μl of reaction mixtures to construct a standard curve for each target species (). In duplex PCR assay, each concentration of both donkey and pork DNA was added to the reaction mixtures to construct the standard curve for the assay ().
Gel Electrophoresis of PCR Products
The amplified fragments were analyzed by electrophoresis in a 3.0% agarose gel prepared with ethidium bromide. The analysis was carried out in 1% TAE buffer (40 mM Tris-acetate, 1 mM EDTA) for 90 min at 80 V. The agarose gel was visualized under UV light and a digital image was recorded using a digital imaging system (Hercules GEL DOC XR, Biorad, USA).
Table 1 Cycle of threshold (Ct) values for each species tested with the species-specific primer-probe sets
RESULTS AND DISCUSSION
Specificity and Sensitivity of Duplex Real-Time PCR System
The specificity of the optimized duplex PCR system was tested for seven livestock species: donkey, pork, horse, cattle, sheep, chicken, and turkey. In this study, it was determined from the specificity test that the duplex PCR system produced similar results with those stated in a previous study[ Citation 12 ] where the simplex PCR system was performed using the same primer and probe sets; namely, the duplex system in this study showed no cross-reaction with any of the non-targeted species because of high specificity of the primer and probe sets used in this study.
The sensitivity and linearity of the duplex real-time PCR technique were compared with those of the simplex PCR technique using 10-fold serial dilutions starting from 100 ng (100%) to 0.001 ng of the genomic DNA obtained from each target species. As shown in , both simplex and duplex real-time PCR systems produced a sensitivity of 0.001 ng (corresponding to 0.001%) DNA for the tested species, donkey and pork. This sensitivity was higher than that reported by Rodriguez et al.,[ Citation 10 ] who determined the detection limit as 0.01 ng using the TaqMan probe system specific to pig species on mitochondrial 12S rRNA. In addition, cycle of threshold (Ct) values were plotted versus log DNA concentrations to construct a standard curve on genomic DNA from each target species. Then, the curve was subjected to linear regression and the magnitudes of the slopes and correlations (R 2) were computed from the raw data to test the system and determine the range of simplex and duplex PCR linearity ( and ). It is clearly seen from and that linear correlations (R 2 > 0.998) were observed over the whole concentration range of each targeted species tested. The slopes (−3.35 and −3.29, ) obtained using simplex PCR and those (−3.23 and −3.25, ) obtained using duplex PCR assay were closer to the theorical value (−3.32), which can be achieved with a PCR efficiency of 100%.
Duplex Real-Time PCR Analysis of Meat Mixtures
Multiplex PCR amplifications are known to decrease sensitivity as compared to one target reaction (simplex PCR). In this study, however, very close levels of detection were achieved by using the duplex PCR technique for each species (0.001%). In other words, it was possible to obtain a detection limit of 0.001% for raw meat and heat-processed meat at 200°C and of 0.01% for heat-processed meat at 120°C for donkey and pork species in beef () by the simultaneous amplification of both targets, namely, that of the fragments of mitochondrial ND2 and ND5 genes in donkey and pork, respectively.
Table 2 Duplex PCR results for raw and heat-processed binary meat mixtures
The success of the obtained results could be attributed to the high sensitivity and specificity level achieved in this study. Furthermore, a linear change was observed in the Ct values corresponding to the DNA concentration of target species in raw and cooked meat samples (). The linearity test parameters for duplex PCR results also revealed that the determination coefficients for raw and cooked samples were seen to change between 0.954 to 0.996, which proved that the developed duplex PCR assay could be easily used to screen for donkey or pork meat adulteration in beef with high sensitivity. One factor contributing to these results was the application of a multi-step dilution method used in the preparation of the binary meat mixtures, which made the identification of the target species possible even at the lowest adulteration level (0.001%). The use of the duplex PCR technique was reported for the detection of various species in different meat sources in the literature. For example, pork was detected in fresh sausages prepared with horse meat, but no level of detection was reported.[ Citation 28 ] Krcmar and Rencova[ Citation 29 ] proposed the duplex PCR technique allowing the detection of 0.01% of pork and chicken meats and bone meal. A multiplex PCR assay was conducted by Matsunaga et al.,[ Citation 30 ] who reported that the identification of cattle, pig, chicken, sheep, goat, and horse meat could be achieved with a detection limit of 0.25 ng DNA for all species. The multiplex real-time PCR technique was used to detect and quantify DNA from duck species,[ Citation 22 ] beef, pork, chicken, turkey, horse meat, mutton, and goat.[ Citation 26 ,Citation 31 ] Finally, Soares et al.[ Citation 15 ] used a duplex PCR assay to quantitatively detect poultry meat adulteration with pork with a detection limit of 0.1% of pork species in poultry meat.
Table 3 Linearity test parameters for duplex PCR results for raw and heat-processed binary meat mixtures
As for the effect of heat treatment on PCR assay, no difference was observed between the Ct values of raw and oven-cooked samples (200°C for 30 min) (), which was consistent with the results in the authors' previous study where the Ct values of raw and cooked samples (180°C for 5 min) were similar to each other. This result could be attributed to the fact that the amplified ND2 and ND5 fragments using the species-specific primers were not affected by the heat treatment because the length of these fragments was so small (shorter than 150 bp) that no amplification problems caused by intense DNA fragmentations could occur in highly processed food products where the possibility of DNA degradation is very high.[ Citation 12 ] Furthermore, it was thought that the DNA structure did not degrade in the cooked samples (200°C) in this study because the inner temperature did not exceed 100°C in the meat products cooked under general atmospheric conditions even if they were cooked at 200°C. On the other hand, the Ct values of cooked samples (120°C) were considerably higher than those of the raw and cooked samples (200°C) (). A possible reason for this could be explained by the fact that the heat application was carried out at 120°C under pressure at 15 psi (autoclaved), not under normal atmospheric conditions. In this way, the applied heat could homogeneously penetrate the meat samples, resulting in higher damage to the DNA structure and leading to increased Ct values ().
When the Ct values of the donkey samples autoclaved at 120°C were compared with those of the pork samples cooked at the same temperature level, the Ct values of donkey were lower than those of pork at each adulteration level (). This indicated that donkey DNA had a higher resistance to heat treatment than that of pork DNA. The reason could be attributed to the fact that an amplified fragment of shorter length might have had higher resistance to the heat treatment. Accordingly, in this study, the lengths of the amplified fragments were 83 and 115 bp in length for donkey and pork, respectively.
Gel Electrophoresis
The quality of amplified DNA fragments from both reference dilutions and raw and heat processed binary meat mixtures was examined by a semi-quantitative approach, which analyzes PCR band intensities using agarose gel electrophoresis. Gel electrophoresis of amplification products indicates the template DNA integrity because alterations in the length of DNA fragments are dependent on the examined material, degree of processing, and DNA extraction method.[ Citation 15 ] In addition, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) technique is used to employ the differences in specific electrophoresis pattern of the expressed protein in each animal species.[ Citation 32 ] In the authors' study, the gel electrophoresis of simplex and duplex PCR products from reference dilutions is shown in , which also indicates the amplification products of donkey and pork species with 83 and 115 bp, respectively. By using the duplex PCR technique, it was possible to clearly separate the amplification products of each species in the same line. The binary reference dilutions showed that as the concentration of the reference dilutions decreased from 100% (corresponding to 100 ng) to 0.01%, band intensities of PCR products generally decreased. The decrement in band intensities indicates that semi-quantitative detection could be possible.
Figure 3 Gel electrophoresis of simplex and duplex PCR products from reference dilutions. M: 600 bp ladder (Qiagen, Gelplot 100, Germany). Lanes D1–D6: Donkey (100, 10, 1, 0.1, 0.01, and 0.001%). Lanes (D + P1)–(D + P6): Donkey–Pork binary mixtures (100, 10, 1, 0.1, 0.01, and 0.001%). Lanes P1–P6: Pork (100, 10, 1, 0.1, 0.01, and 0.001%).

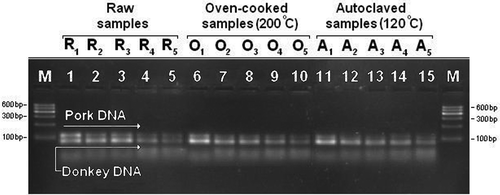
The success of the developed duplex real-time PCR assay for raw and cooked binary meat mixtures was examined using agarose gel electrophoresis. By analyzing the band intensities of PCR products using image analysis software, it was observed that the fluorescence intensities of the duplex PCR bands were correlated with pork and donkey meat percentages (). The correlation was seen in both raw and heat processed (autoclaved and oven cooked) samples, indicating that the band intensities decreased as the pork and donkey meat percentages decreased from 100% (corresponding to 100 ng) to 0.01%. Gel electrophoresis analysis revealed that the band intensities obtained for autoclaved pork samples were lower than those obtained for autoclaved donkey meat samples (). As mentioned above, similar trends were observed in the Ct values of the autoclaved pork and donkey meat samples. In other words, the parallel results were obtained with duplex real-time PCR assay and gel electrophoresis.
Figure 4 Gel electrophoresis of duplex PCR products from raw and heat-processed binary donkey/pork meat mixtures amplified with 40 cycles. M: 600 bp ladder (Qiagen, Gelplot 100, Germany). Lanes R1–R5: Raw donkey-pork binary mixtures in beef (50, 10, 1, 0.1, and 0.01%). Lanes O1–O5: Oven-cooked (200°C) donkey–pork binary mixtures (50, 10, 1, 0.1, and 0.01%). Lanes A1–A5: Autoclaved (120°C) donkey–pork binary mixtures (50, 10, 1, 0.1, 0.01%).
CONCLUSIONS
TaqMan-based duplex real-time PCR assays were developed in this study for the detection of donkey and pork species adulteration in beef with a sensitivity of 0.001%. The duplex PCR technique proved to be a reliable, specific, highly sensitive, and readily applicable tool for the determination of adulteration levels in both raw and cooked meat samples processed under different cooking conditions. In this respect, the TaqMan-based duplex real-time PCR assay can be recommended to determine even the accidental or intentional adulterations in highly processed raw and cooked meat products.
REFERENCES
- Asensio , L. , Gonzalez , I. , Garcia , T. and Martin , R. 2008 . Determination of food authenticity by enzyme-linked immunosorbent assay (ELISA) . Food Control , 19 : 1 – 8 .
- Cota-Rivas , M. and Vallejo-Cordoba , B.V. 1997 . Capillary electrophoresis for meat species differentiation . Journal of Capillary Electrophoresis , 4 : 195 – 199 .
- Andrews , C.D. , Berger , R.G. , Mageau , R.P. , Schwab , B. and Jhonston , R.W. 1992 . Detection of the beef, sheep, deer, and horse meat in cooked meat products by enzyme-linked immunosorbent assay . Journal of AOAC International , 75 : 572 – 576 .
- Chen , F.C. and Hsieh , Y.H. 2000 . Detection of pork in heat-processed meat products by monoclonal antibody-based ELISA . Journal of AOAC International , 83 : 79 – 85 .
- Skarpeid , H. J. , Kvaal , K. and Hildrum , K. I. 1998 . Identification of animal species in ground meat mixtures by multivariate analysis of isoelectric focusing protein profiles . Electrophoresis , 18 : 3103 – 3109 .
- Chikuni , K. , Tabata , T. , Kosigiyama , M. and Monma , M. 1994 . Polymerase chain reaction assay for detection of sheep and goat meats . Meat Science , 37 : 337 – 345 .
- Kesmen , Z. , Şahin , F. and Yetim , H. 2007 . PCR assay for the identification of animal species in cooked sausages . Meat Science , 77 : 649 – 653 .
- Mafra , I. , Ferreira , I.M. and Oliveira , M.B. 2008 . Food authentication by PCR-based methods . European Food Research and Technology , 227 : 649 – 665 .
- Hird , H. , Goodier , R. and Hill , M. 2003 . Rapid detection of chicken and turkey in heated meat products using the polymerase chain reaction followed by amplicon visualisation with vistra green . Meat Science , 65 : 1117 – 1123 .
- Rodriguez , M. A. , Garcia , T. , Gonzalez , I. , Hernandez , P.E. and Martin . 2005 . R. TaqMan real-time PCR for detection and quantitation of pork in meat mixtures . Meat Science , 70 : 113 – 120 .
- Guoli , Z. , Mingguang , Z. , Zhijiang , Z. , Hongsheng , O. and Qiang , L. 1999 . Establishment and application of a polymerase chain reaction for the identification of beef . Meat Science , 51 : 233 – 236 .
- Kesmen , Z. , Gulluce , A. , Sahin , F. and Yetim , H. 2009 . Identification of meat species by TaqMan-based real-time PCR assay . Meat Science , 82 : 444 – 449 .
- Fajardo , V. , Gonzales , I. , Martin , I. , Rojas , M. , Hernandez , P.E. , Garcia , T. and Real-time , Martín, R. 2008 . PCR for detection and quantification of red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus) in meat mixtures . Meat Science , 79 : 289 – 298 .
- Mafra , I. , Silva , S.A. , Moreira , E.J.M.O. , Ferreira da Silva , C.S. and Oliveira , M.B.P.P. 2008 . Comparative study of DNA extraction methods for soybean derived food products . Food Control , 19 : 1183 – 1190 .
- Soares , S. , Amaral , J.S. , Mafra , I. and Oliveira , M.B.P.P. 2010 . Quantitative detection of poultry meat adulteration with pork by a duplex PCR assay . Meat Science , 85 : 531 – 536 .
- Lopez-Andreo , M. , Garrido-Pertierra , A. and Puyet , A. 2006 . Evaluation of postpolymerase chain reaction melting temperature analysis for meat species identification in mixed DNA samples . Journal of Agricultural and Food Chemistry , 54 : 7973 – 7978 .
- Walker , J.A. , Hughes , D.A. , Anders , B.A. , Sinha , J. , Shewale , S.K. and Batzer , M.A. 2003 . Quantitative intra-short interspersed element PCR for species-specific DNA identification . Analytical Biochemistry , 316 : 259 – 269 .
- Brodmann , P.D. and Moor , D. 2003 . Sensitive and semi-quantitative TaqManTM realtime polymerase chain reaction systems for the detection of beef (Bos taurus) and the detection of the family Mammalia in food and feed . Meat Science , 65 : 599 – 607 .
- Chisholm , J. , Conyers , C. , Booth , C. , Lawley , W. and Hird , H. 2005 . The detection of horse and donkey using real-time PCR . Meat Science , 70 : 727 – 732 .
- Dooley , J.J. , Paine , K.E. , Garrett , S.D. and Brown , H.M. 2004 . Detection of meat species using TaqMan real-time PCR assays . Meat Science , 68 : 431 – 438 .
- Hird , H. , Goodier , R. , Schneede , K. , Boltz , C. , Chisholm , J. , Lloyd , J. and Popping , B. 2004 . Truncation of oligonucleotide primers confers specificity on real time-PCR assays for food authentication . Food Additives and Contaminants , 21 : 1035 – 1040 .
- Hird , H. , Chisholm , J. and Brown , J. 2005 . The detection of commercial duck species in food using a single probe-multiple species-specific primer real-time PCR assay . European Food Research and Technology , 221 : 559 – 563 .
- Krcmar , P. and Rencova , E. 2005 . Quantitative detection of species-specific DNA in feedstuffs and fish meals . Journal of Food Protection , 68 : 1217 – 1221 .
- Zhang , C.L. , Fowler , M.R. , Scott , N.W. , Lawson , G. and Slater , A. A . 2007 . TaqMan real-time PCR system for the identification and quantification of bovine DNA in meats, milks and cheeses . Food Control , 18 : 1149 – 1158 .
- Holland , P.M. , Abramson , R.D. , Watson , R. and Gelfand , D.H. 1991 . Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Termus aquaticus DNA polymerase . Proceedings of the National Academy of Sciences , 88 : 7276 – 7280 .
- Köppel , R. , Ruf , J. , Zimmerli , F. and Breitenmoser , A. 2008 . Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey . European Food Research and Technology , 227 : 1199 – 1203 .
- Heptinstall , J. and Rapley , R. 2002 . “ Spectrophotometric analysis of nucleic acids ” . In The Nucleic Acid Protocols Handbook , Edited by: Rapley , R. 57 – 60 . Totowa , NJ : Humana Press .
- Di Pinto , A. , Forte , V.T. , Conversano , M.C. and Tantillo , G.M. 2005 . Duplex polymerase chain reaction for detection of pork meat in horse meat fresh sausages from Italian retail sources . Food Control , 16 : 391 – 394 .
- Krcmar , P. and Rencova , E. 2003 . Identification of species-specific DNA in feedstuffs . Journal of Agricultural and Food Chemistry , 51 : 7655 – 7658 .
- Matsunaga , T. , Chikuni , K. , Tanabe , R. , Muoya , S. , Shibata , K. , Yamada , J. and Shinmura , Y. 1999 . A quick and simple method for the identification of meat species and meat products by PCR assay . Meat Science , 51 : 143 – 148 .
- Köppel , R. , Zimmerli , F. and Breitenmoser , A. 2009 . Heptaplex real-time PCR for the identification and quantification of DNA from beef, pork, chicken, turkey, horse meat, sheep (mutton) and goat . European Food Research and Technology , 230 : 125 – 133 .
- Che Man , Y.B. , Mustafa , S. , Mokhtar , N.F.K. , Nordina , N. and Sazilia , A.Q. “ Porcine-specific polymerase chain reaction assay based on mitochondrial d-loop gene for identification of pork in raw meat ” . In International Journal of Food Properties doi: 10.1080/10942911003754692