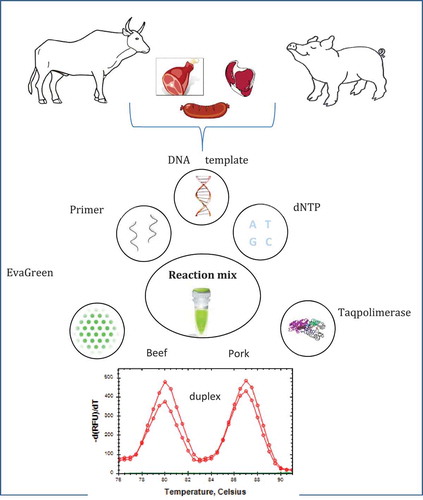
Abstract
In this study, a rapid, specific, and low-cost duplex-detection technique of pork and beef was developed by a real-time polimerase chain reaction assay based on fluorescence. Deoxyribonucleic acid was extracted from variable mixtures of pork and beef in sausage and industrial products to develop the duplex assay using the GIDAGEN® Multi-Fast DNA Isolation Kit. Identification of genomes was accomplished in the same tube by their distinctive melting peak, which was 87.5°C for pork and 80.5°C for beef, respectively. The detection limit of the method was 0.01 ng/µL deoxyribonucleic acidor 0.001% target pork and beef in sausage. The results showed that the intercalating dye based duplex real-time polimerase chain reaction is a potentially sensitive, reliable, and practical assay for the detection of meat species adulterated with beef and pork.
Introduction
The identity of meat species is not always readily apparent and accurate. In recent years, species determination of meat and meat products has gained increased attention. In most countries, food manufacturers are prone to use cheaper meats instead of expensive ones. In addition, the impure labeling of meat products has become common in the meat industry and raised a number of concerns regarding health, diet, religion, and lifestyle issues.[Citation1,Citation2] Food labeling regulations require that the species of meat in food products have to be accurately declared to the consumer.
The authentication of meat and meat products can be identified by different analytical methodologies including chromatography[Citation3] spectroscopy,[Citation4] mass spectrometry,[Citation5] electronic spin resonance,[Citation6] microscopy, enzymatic assays, and polymerase chain reaction.[Citation7,Citation8] The stability of deoxyribonucleic acid (DNA) under high temperatures, pressures, and chemical treatments used during processing ascertains the specificity and reliability of DNA-based methods.[Citation2] Therefore, the techniques of DNA identification have an enormous potential for diagnostics, forensics, and food analysis.[Citation9] The agarose gel-based multiplex polimerase chain reaction (PCR) assay was used by various workers for simplex, duplex, or multiplex identification of multiple meat species in single PCR reaction employing single or multiple primer pairs. The detection limits by multiplex assay were reported to be between 0.002 and 3.6% by various workers.[Citation10–Citation16] Computer imaging-based real-time PCR was employed by various researchers for simultaneous determination of multiple meat species in single PCR reaction employing multiple primer pairs.[Citation17] Real time PCR has attracted great attention in molecular techniques for the detection of the origin of meat,[Citation18] allergen content of foods,[Citation19] and pathogen bacteria in meats[Citation12] due to its sensitivity, simplicity, and reproducibility. The fluorescent based methods used in real-time PCR can be classified into two categories: probe-based such as TaqMan[Citation20] and DNA intercalating dyes, such as the SYBR Green and EvaGreen.[Citation21] Although probe-based ones are sequence-specific, they are more expensive,[Citation23] time- and labor-intensive, and much more difficult to design and optimize. In addition to these, probe-based (TaqMan) real-time PCR is destitute of the ability to perform a melt-curve analysis to check the specificity of the amplification reaction. Alternatively, EvaGreen binds into the minor groove of double-stranded DNA in a sequence-independent way. Moreover, it provides a flexible method without the need for individual probe design and complex optimization steps.[Citation24] Economically, the use of EvaGreen Green is cheaper than probe-based methods.[Citation25] It is much less inhibitory to PCR than SYBR® Green I dye and, consequently, can be used at higher (saturating) concentrations to enable consistent and superior discrimination of PCR-amplified DNA[Citation26,Citation27] with melting curve analysis.
In our previous study, SYBR Green intercalating dye based duplex real time PCR technique was developed for the identification of ruminant and poultry meat origins in foodstuff.[Citation25] In this article, we describe an EvaGreen intercalating dye based duplex assay, which appears to be a promising tool for identification of beef and pork genes in meat and meat products ().
Materials and Methods
Preparation of Meat Samples
Reference raw meat samples, beef (Bostaurus) and pork (Susscrofa), were purchased from local suppliers. For the applicability and sensitivity of the assay, reference samples (i.e., pork and beef) supplied as raw meat were mixed to form mixtures (50 g).The binary homogeneous mixtures were diluted to obtain 0.00001, 0.0001, 0.001, 0.01, 0.1, 1, and 10 % (wt/wt) pork and beef species meat in the sausages prepared from chicken meat, soybean protein, and additive materials (such as spices) by Meat Products Industry (Danet, Afyon, Turkey). Samples of raw meats (n = 5) and two types of commercial sausages, which were 100% beef (sausage 1 not containing soybean and poultry; n = 5), 100% chicken (sausage 2 containing poultry and soybean protein; n = 5) were purchased randomly from the local companies for the application of assay. The obtained samples were directly transported to the Molecular Bioengineering and Genetic Research Laboratory of Çanakkale 18 Mart University. All samples were stored at −20°C until the extraction of DNA in order to prevent the enzymatic degradation of DNA.
DNA Extraction
DNA was extracted from meat and meat products with Multi-Fast DNA Isolation Kit (GIDAGEN, İstanbul, Turkey). According to its manual, 100 mg of homogenized meat and sausages was transferred in a 2 mL centrifuge tube. Five hundred µL buffer Multi Lysis (ML) and 20 µL Proteinase K was added into the tube and mixed by the vortex for 15–20 s. Then, tubes were incubated samples at 65°C for 5 min in the block heater. The vortex was applied to tubes for 30–45 s after incubation and centrifuged at the speed of least 13,300 rpm (17,000 × g) for 5 min. A liquid portion of 230 µL was transferred in a new 2 mL tube and added 500 µL buffer Multi Binding (MB) to the tube and mixed again thoroughly by the vortex. The mixture (730 µL) was transferred in the spin column with 2 mL collection tube and centrifuged at 9000 rpm (7800 × g) for 1 min. Then, the aqueous phase was discarded and spin column was placed the in same collection tube. Buffer Purification (P; 500 µL) of was added into the spin and centrifuged at 13,300 rpm (17,000 × g) for 3 min. The spin column was placed into a new 1.5 mL/2 mL centrifuge tube and added 75 µL of buffer Elution (E) into the spin column. The tube was kept at room temperature for 3 min and then centrifuged at 10,000 rpm (9600 × g) for 1 min. Again 75 µL of buffer E was added in the spin column, and then centrifuged at 10,000 rpm (9600 × g) for 1 min. After the concentration of DNA was measured, the DNA solutions (150 µL) were stored at –20°C in prior to use.
Nucleic Acid Quantification
Infinite® 200 PRO (Tecan, Switzerland) was used to determine the concentration of DNA in the solution. The samples were exposed to ultraviolet light at 260 and 280 nm. We used 260:280 to calculate the qualification of nucleic acids by the following formula: DNA concentration = OD260 × extinction coefficient (50 µg/mL) × dilution factor.
Primers
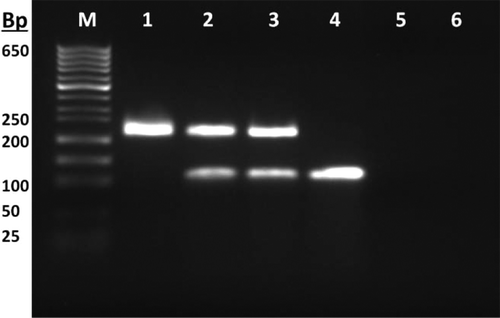
Beef (Bos taurus) and pork (Sus sacrofa) primers published by Rodríguez and others (2004)[Citation21] and Köppel and others (2009)[Citation22] were used, respectively. All primers were synthesized by the Metabion (Steinkirchen, Germany) company. A software-tool called BLAST (Basic Local Alignment Search Tool) of NCBI (National Center for Biotechnology Information) was used to check the specificity of each primer. The primers for species-specific amplification of beef genomic DNA were 5’-CTA GAG GAG CCT GTT CTA TAA TCG ATA A-3’ (forward) and 5’-TGG TTT CAT AAT AAC TTT CGC GCT-3’ (reverse). The primers for species-specific amplification of pork genomic DNA were 5’-CGA CGA GGC TGC CGT AAA GG -3’(forward) and 5’-TGC AAG GAA CAC GGC TAA GTG -3’ (reverse). The sizes of the expected beef and pork amplicons were 223 and 111 bp, respectively.
Specificity and Sensitivity
The specificity of each species specific primer was controlled by amplification of 100 ng of pork (Sus sacrofa), beef (Bos taurus), sheep (Ovis aries), goat (Capra hircus), chicken (Gallus gallus), and soybean (Glycine max) genomic DNA. To determine the absolute detection limit of EvaGreen based duplex real-time PCR, a mixture (1/1) of beef and pork DNA was adjusted to200 ng/μL and subsequently serially diluted by 10-fold increment (yielding solutions of 200, 20, 2, 0.2, 0.02, 0.002, 0.0002, and 0.00002 ng/μL. DNA formed binary homogeneous mixtures were amplified to identify the detection limit of EvaGreen based duplex real-time PCR assay.
Optimization of the Primer Concentration for Duplex Real-Time PCR
The optimization of primer quantity in the PCR mix was carried out by mixing primers at variable quantities so that both amplicons were produced with high efficiency in the same reaction by EvaGreen based duplex real-time PCR. In this research, the assay was developed with equal concentrations of beef and pork primers.
Evagreen-Based Duplex Real-Time PCR Protocol
The PCR amplification was performed in a final volume of 20 µL containing 4 µL of EvaGreenq PCR Mix Plus (Solis BioDyne, Estonia), 10 pmol of cattle and pork primers, and 200 ng DNA template extracted from reference mixtures. The amplification was performed in a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). After an initial heat denaturation step at 94°C for 10 min, 35 cycles were programmed as follows: 94°C for 15 s, 58°C for 20 s, and 72°C for 20 s. After that, a melting curve analysis was programmed in such a way that its ramp formed from 72 to 95°C by raising 1°C each step.
Identification of Species by Melting Curve Analysis
At the end of each reaction, melting curve analysis tools of the CFX Manager SoftwareVersion 3.1 (Bio-Rad, Hercules, CA, USA) was used to identify species-specific mp values of the amplified region of the template DNA that belong to reference meat samples.
Gel Electrophoresis
PCR products were analyzed by electrophoresis on 2% agarose gel with ethidium bromide (0.2 mg/mL) run in 0.5X TBE buffer for 70 min at 110 V. Images were recorded with a digital image under UV light.
Results and discussion
DNA Extraction
The spectrophotometer results showed that extracted DNA was suitable for PCR amplification. The DNA extraction method was considered satisfactory and able to remove PCR inhibitors, which could interfere with PCR reaction. The purity and yields of the total DNA extract obtained from the reference meats and commercial sausages were high (purity = A260/A280 and 260/230 ratio ranged between 1.8 and 2.0 and yield = 100–120 ng/μL).
Design of Evagreen Based Duplex Real-Time PCR for Analysis of Beef and Pork
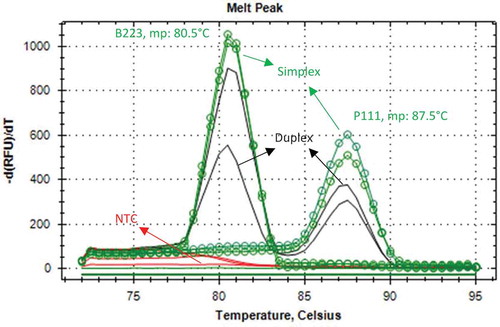
The primers specific to beef and pork were combined for the duplex reaction. The templates were amplified repeatedly in the real-time PCR followed by a melting curve analysis using EvaGreen and CFX96 Touch™ Real-Time PCR Detection System. The accumulation of amplicons in the same reaction was demonstrated in representing the changes in fluorescence as a function of time –d(RFU)/dT versus the temperature of the reaction products. Beef and pork amplicons were easily distinguished through specific mp values due to the different base (especially GC) compositions of two amplicons. EvaGreen based real-time PCR resulted in a single curve with two peaks for each species as shown in . These peaks were formed at a specific location on the temperature axis at 87.5 and 80.5°C for the duplex reaction of beef and pork, respectively. Simplex and duplex PCR products were run on an agarose gel stained with ethidium bromide for crosschecking. Agarose gel electrophoresis of the PCR products showed that beef and pork samples produced clear bands of the expected size of 223 and 111 bp, respectively ().
Sensitivity of the Duplex Real-Time PCR Assay
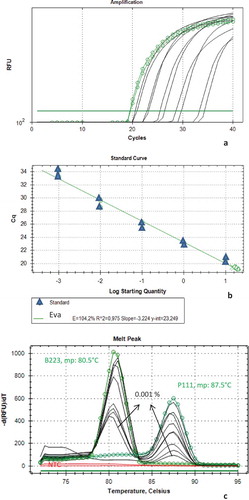
The analytical sensitivity of EvaGreen based real-time PCR assay was determined by using DNAs extracted from the dilution series of homogeneous mixture(reference sausage), which contained pork and beef meats (). The alignment of the experimental points in the standard curve was verified. The curve slope, amplification efficiency (E ¼ [10-1/slope] – 1) and RCitation2 were also calculated using the CFX Manager Software. The amplification plot () generated a slope of –3.224 corresponding to a 104.2% efficiency of the PCR assay. A linear relationship between the input DNA and the Ct values with a regression coefficient (RCitation2) of 0.975with 23.49 as the value of y-intercept was obtained for amplification representing two meat samples. Duplex real-time PCR assay sensitivity was 0.001% for the specific duplex detection of beef and pork in the reference sausage (). The absolute detection limit of EvaGreen based duplex real-time PCR was 0.0002 ng/µL for each of beef and pork (figure not shown).
Specificity of Duplex Real-Time PCR
The specificity results of the beef and pork primers showed no cross-reaction with any of the non-target species after optimization. The specificity of the duplex assay was established by carrying out melting curve analysis. The duplex real-time PCR assay was specific for each species through the use of melting peaks. Although beef and pork primers in negative template control (ntc) rarely produced dimer or unspecific amplicons appeared after 33rd cycle of real-time PCR, there was no significant relationship between the mp values of unspecific amplicon or primer dimer and the mp values of duplex amplicons of beef and pork (). All amplicons and primer dimmers appearing after 33rd cycle were ignored due to the possibility of interference with specific amplicons and the cross-reactivity of beef primers with non-target species.
TABLE 1 Mp and average Cq values of some commercial meat and meat products
Application of Intercalating Dye Based Duplex Assay to Commercial Sausages and Raw Meats
Duplex real-time PCR was applied to the samples of raw meats and two types of commercial sausages in duplicate. The samples of raw meats and commercial sausages 1 indicated that 100% beef generated positive results of beef, but none of them yielded pork amplicons. The samples of commercial sausages 2 indicated 100% chicken and did not yield positive results for pork but two of these sausages yielded positive results for beef with values of Cq (cycle quantification) 30.2 and 29.3 as shown in . The control sample (Cq: 20.6) representing 10% of reference mixture DNA was assigned, then the relative quantity of two of aforementioned samples was calculated with this equation: Relative quantity = E Mean Cq of Controls – Mean Cq of Sample; CFX Manager™ Software Help Data Analysis Details). According to this equation, the mean Cq values of 30.2 and 29.3 indicated that two of the commercial sausages contained beef by 0.01 and 0.02%, respectively. The quantity of the samples was also calculated by a different equation: Cq = slope (log) + y-intercept and provided 0.083 and 0.15%. In addition, the difference between the Ct value of tested samples (unknown) and the Ct value of the reference (ptc) indicates the percentage of tested (unknown) samples without constructing a standard curve in Şakalar quantification table (SQT-DNA).[Citation27] The concentration of DNA amplicon on 29.3rd cycle has been known around 0.04324% in SQT-DNA when Cq (Ct) value of the 20.6th cycle is assigned as the positive control. Unexpected results might be associated with contamination (it indicates unhygienic conditions) or an unintentional contaminant but not intentional use (fraud) since the level of quantity was not high enough to be profitable.
Duplex real-time PCR fragments were detected by melting curve analysis. The melting temperature (tm) which is specific for each amplicon, is the temperature at which 50% of the DNA amplicon is in a double-stranded configuration. The tm depends on various factors including the amplicon length and the nucleotide sequence. A common problem with intercalating dye based real-time PCR is the presence of a non-predicted amplicon product. The most frequent such products are derived from primer dimers, which are most evident in samples with little specific target template. In this study, primer dimer did not influence the results considerably due to a good primer, which set results in a single product, distinguished by producing a single melting curve peak.[Citation23] Pork and beef amplicons can easily be distinguished by specific mp values due to different lengths and GC compositions of two target regions. Melting curve analysis revealed that the beef melting curve was a Tm of 80.5°C, whereas the pork melting curve was a Tm of 87.5°C ().
CONCLUSIONS
EvaGreen-based duplex real-time PCR assays were developed in this study for the detection of beef and pork adulteration in meat and meat products with a sensitivity of 0.001%. The results of the present study implicated that duplex methodology can be used as a sensitive tool with high sensitivity and specificity for the detection of beef and pork in meat and meat products using EvaGreen based duplex real-time PCR. The applicability of the assay was demonstrated by the laboratory validation using binary meat mixtures. Future studies should concentrate on describing the detection of more than two species in the same reaction mixture.
Acknowledgments
The authors would like to thank Food Gene Analytic Technology and Designing (GIDAGEN) and The Scientific and Technological Research Council of Turkey (TUBITAK) that has provided the equipment of their laboratory in this study (Project Grant No. 113Z831).
REFERENCES
- Asensio, L.; Gonzalez, I.; Garcia, T.; Martin, R. Determination of Food Authenticity by Enzyme-Linked Immune Sorbent Assay (ELISA). Food Control 2008, 19, 1–8.
- Sakaridis, I.; Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. A Fast and Accurate Method for Controlling the Correct Labeling of Products Containing Buffalo Meat Using High Resolution Melting (HRM) Analysis. Meat Science 2013, 94, 84–88.
- Hartwig, M.; Hartmann, S.; Steinhart, H. Physiological Quantities of Naturally Occurring Steroid Hormones (Androgens and Progestogens), Precursors and Metabolites in Beef of Differing Sexual Origin. Z Lebensm Unters Forsch 1997, 205, 5–10.
- Huck-Pezzei, V.A.; Seitz, I.; Karer, R.; Schmutzler, M.; Benedictis, L.D.; Wild, B.; Huck, C.W. Alps Food Authentication, Typicality, and Intrinsic Quality by Near Infrared Spectroscopy. Food Research International 2014, 62, 984–990.
- Nurjuliana, M.; Che Man, Y.B.; Hashim, D.M.; Mohamed, A.K.S. Rapid Identification of Pork for Halal Authentication Using the Electronic Nose and Gas Chromatography Mass Spectrometer with Headspace Analyzer. Meat Science 2011, 88, 638–644.
- Chawla, S.P.; Thomas, P. Identification of Irradiated Meat Using Electron Spin Resonance Spectroscopy: Results of Blind Trials. International Journal of Food Science and Technology 2004, 39, 653–660.
- Soares, S.; Amaral, J.S.; Mafra, I.; Oliveira, M.B. Quantitative Detection of Poultry Meat Adulteration with Pork by a Duplex PCR Assay. Meat Science 2010, 85, 531–536.
- Kitpipit, T.; Sittichan, K.; Thanakiatkrai, P. Direct-Multiplex PCR Assay for Meat Species Identification in Food Products. Food Chemistry 2014, 163, 77–82.
- Lopez-Andreo, M.; Garrido-Pertierra, A.; Puyet, A. Evaluation of Post-Polymerase Chain Reaction Melting Temperature Analysis for Meat Species Identification in Mixed DNA Samples. Journal of Agricultural and Food Chemistry 2006, 54, 7973–7978.
- Behrens, M.; Unthan, M.; Bronkmann, Y.; Buchholz, R.; Lotus, N. Identification of Animal Species in Heated and Complex Meat Products Using Specific PCR Reactions. Fleschwirtschaft 1999, 79, 97–100.
- Matsunaga, T.; Chikuni, K.; Tanabe, R.; Muroya, S.; Shibata, K.; Yamada J.; Shinmura, Y. A Quick and Simple Method for the Identification of Meat Species and Meat Products by PCR Assay. Meat Science 1999, 51, 143–148.
- Dalmasso, A.; Fontanella, E.; Piatti, P.; Civera, T.; Rosati, S.; Bottero, M.T. A Multiplex PCR Assay for the Identification of Animal Species in Feedstuffs. Molecular and Cellular Probes 2004, 18, 81–87.
- Şakalar, E.; Abasiyanik, M.F. Qualitative Analysis of Meat and Meat Products by Multiplex Polymerase Chain Reaction (PCR) Technique. African Journal of Biotechnology 2011, 10, 9379–9386.
- Rea, S.; Chikuni, K.; Branciari, R.; Sangamayya, S.R.; Ranucci, D.; Avellini, P. Use of Duplex Polymerase Chain Reaction (Duplex-PCR) Technique to Identify Bovine and Water Buffalo Milk Used in Making Mozzarella Cheese. Journal of Dairy Research 2001, 68, 689–698.
- CheMan, Y.B.; Mustafa, S.; Mokhtar, N.F.; Nordin, R.; Sazili, A.Q. Porcine-Specific Polymerase Chain Reaction Assay Based on Mitochondrial D-Loop Gene for Identification of Pork in Raw Meat. International Journal of Food Properties 2012, 15, 134–144.
- Kumar, A.; Kumar, R.R.; Sharma, B.D.; Gokulakrishnan, P.; Mendiratta, S.K.; Sharma, D. Identification of Species Origin of Meat and Meat Products on the DNA Basis: A Review. Critical Reviews in Food Science and Nutrition 2015, 55, 1340–1351.
- Kesmen, Z.; Güllüce A.; Yilmaz, M.T.; Yetiman, A.E.; Yetim, H. Taqman-Based Duplex Real-Time Polymerase Chain Reaction Approach for the Detection and Quantification of Donkey and Pork Adulterations in Raw and Heat-Processed Meats. International Journal of Food Properties 2014, 17, 629–638.
- Langen, M.; Peters, U.; Körner, U.; Gissel, C.; Stanislawski, D.; Klein, G. Semi Quantitative Detection of Male Pork Tissue in Meat and Meat Products by PCR. Meat Science 2010, 86, 821–824.
- Gomez, G.A.M.; Brohee, M.; Silva, E.A.; van Hengel, A.J.; Chassaigne, H. Development of a Real-Time PCR Method for the Simultaneous Detection of Soya and Lupine Mitochondrial DNA As Markers for the Presence of Allergens in Processed Food. Food Chemistry 2011, 127, 834–841.
- Malorny, B.; Bunge, C.; Helmuth, R. A Real-Time PCR for the Detection of Salmonella Enteritisin Poultry Meat and Consumption Eggs. Journal of Microbiological Methods 2007, 70, 245–251.
- Rodríguez, M.A.; Garcia, T.; Gonzalez, I.; Asensio, L.; Hernandez, P.E.; Martín, R. PCR Identification of Beef, Sheep, Goat, and Pork in Raw and Heat-Treated Meat Mixtures. Journal of Food Protection 2004, 1, 4–214.
- Köppel, R.; Zimmerli, F.; Breitenmoser, A. Heptaplex real-Time PCR for the Identification and Quantification of DNA from Beef, Pork, Chicken, Turkey, Horse Meat, Sheep (Mutton) and Goat. European Food Research and Technology. 2009, 230, 125–133.
- Fajardo, V.; Gonzalez, I.; Martin, I.; Rojas, M.; Hernandez, P.E.; Garcia, T.; Martin, R. Real-Time PCR for Detection and Quantification of Red Deer (Cervus elaphus), Fallow Deer (Dama dama), and Roe Deer (Capreolus capreolus) in meat mixtures. Meat Science 2008, 79, 289–298.
- Martin, I.; Garcia, T.; Fajardo, V.; Rojas, M.; Pegels, N.; Hernandez, P.E.; Gonzalez, I.; Martin, R. SYBR-Green Real-Time PCR Approach for the Detection and Quantification Of Pig DNA in Feedstuffs. Meat Science 2009, 82, 252–259.
- Terzi, V.; Infascelli, F.; Tudisco, R.; Russo, G.; Stanca, A.M.; Faccioli, P. Quantitative detection of Secale Cereale by Real-Time PCR Amplification. LWT-Food Science and Technology 2004, 37, 239–246.
- Şakalar, E.; Abasıyanık, M.F. The Development of Duplex Real-Time PCR Based on SYBR Green Florescence for Rapid Identification of Ruminant and Poultry Origins in Foodstuff. Food Chemistry 2012, 130, 1050–1054.
- Mao, F.; Leung W.Y.; Xin, X. Characterization of Eva Green and the Implication of Its Physicochemical Properties for qPCR Applications. BMC Biotechnology 2007, 7, 76.
- Şakalar, E. The Practical Analysis of Food: The Development of Şakalar Quantification Table of DNA (SQT-DNA). Food Chemistry 2013, 141, 718–722.