Abstract
The identifications of species in meat products have created interests since these foods became the target of forgery and fraud in the market. The presence of pork in food products is not allowed for the Muslim community. Hence, an analysis is necessary to detect the presence of pork in processed meat products, such as in dendeng (dried meat) product. Real time polymerase chain reaction using mitochondrial displacement loop686 and cytochrome b (cytb) gene primers was used to identify specific pork DNA among other four types of DNA species; namely beef, chicken, goat, and horse. This method was also used to identify pork DNA in the laboratory processed pork-beef dendeng as well as commercial dendeng from market. The results showed that real time polymerase chain reaction using displacement loop686 and cytb gene primers were able specifically to distinguish between pork DNA and the other species. The lowest concentration of 0.5% of pork DNA in a mixture of pork-beef processed products of dendeng was able to be detected by both primers with the product amplification of 114 and 134 bp (base pair) for the displacement loop686 and 149 bp for cytb gene, respectively. High sensitivity was also obtained when both primers were applied with the lowest detection limit of 5 pg/µL pork DNA. The results of the six commercial dendeng amplification using both primers showed no amplified products present, meaning that these products do not contain porcine DNA.
INTRODUCTION
Beef based food product has usually been adulterated using other meat such as pork due to the price difference between beef and pork. One of the food subjected to adulteration target is dendeng (Indonesia dried meat). The National Food Agency of Republic of Indonesia has announced that 5 out of 35 brand products of beef dendeng positively contained pork meat.[Citation1] This is serious issue because Indonesia is the largest Muslim country, and Muslims are not allowed to consume any products containing pork meat.[Citation2,Citation3] Therefore, the development of analytical method for pork detection is really needed.
Various analytical methods to detect the presence of a mixture of pork have been carried out including the use of engineering Electronic Nose,[Citation4] Fourier Transform infrared (FT-IR) spectroscopyfor analysis of pig fat content in beef meatballs,[Citation5,Citation6] enzyme-linked immunosorbent assay (ELISA) for analysis of pork in meat products which have been processed by heating,[Citation7] and polymerase chain reaction (PCR) for the identification of pork DNA in sausage products, sausage wrappers, bread, and biscuits using 12S ribosomal RNA specific targets (rRNA).[Citation8] The detection of pork contamination in the meatballs using restriction fragment length polymorphism (RFLP)-PCR,[Citation9] and in the raw meat using mitochondrial displacement loop (D-Loop) primers[Citation10] have been performed. Currently, Kesmen et al.[Citation11] have developed Taqman based duplex Real-Time PCR (RT-PCR) approach for the detection and quantification of donkey and pork adulterations in raw and heat-processed meats.
RT-PCR, which based on detecting pork DNA, becomes the method of choice for routine analysis, due to the sensitivity and specificity. In this study, mitochondrial DNA (mtDNA) was chosen as the target for amplification. Since mtDNA has a high copy number (approximately about 100–10,000 per cell), so that it can be used for the analysis of samples with a very limited amount of DNA, and the primer targeted to amplify mtDNA was used to detect and quantify the pork DNA in 17 samples product of commercial processed poultry meat.[Citation12] The primer targeted on the region of D-Loop, D-Loop686, was already designed by Himawati.[Citation13] The used primer was able to specifically amplify pork DNA on beef meatball samples. The primer based on the other region of mitochondria DNA, namely cytochrome b (cytb) was also been developed. This primer showed high specificity for pork identification with the amplification product of 149bp.[Citation14]
In this study, the primer targeted on the D-Loop686 and cytb gene were used for pork identification among four other meats, namely beef, chicken, goat, and horse, as well as laboratory prepared beef dendeng containing various concentration of pork. Furthermore, the two primers were used for analysis the commercial dendeng obtained from several supermarkets and traditional markets in Yogyakarta, Indonesia.
MATERIALS AND METHODS
Preparation of Pork-Beef Dendeng and DNA Extraction
Dendeng was prepared prepared based on a formula described by Purnomo and Rahadiyan.[Citation15] Both raw pork and beef were cleaned, sliced, and ground individually. Forty grams of grounded beef and pork was mixed with the pork concentrations of 0.5, 1, 3, 5, 10% wt/wt in beef. Furthermore, the mixed meat was added with 10 g of spice paste (palm sugar 15%, salt 2%, coriander 4.6%, garlic 0.6%, galangal 0.2%, tamarind 0.6%) and stored for 12 h at –20oC. Pure beef dendeng was prepared as a negative control and pure pork dendeng as positive control. This mixture was spread to form of thin sheets, and dried at 70oC for 3 h.[Citation16]
For DNA isolation, all samples, raw meats (pork, beef, chicken, goat, and horse) and dendengs were grounded. Two hundred fifty milligrams of samples were transferred into 2 mL Eppendorf tube and DNA was extracted using 1 mL of TNE lysis buffer (50 mM TrisHCl pH 8, 100 m MEDTA, 100 mM NaCl and 1% SDS) and 15 µL of proteinaseK (20 mg/mL). This mixture was incubated at 55oC for 1 h, followed by centrifugation. The supernatant was extracted using phenol-chloroform followed by DNA precipitation in two times volume of absolute ethanol. The DNA pellet was then washed using 70% ethanol and dissolved in TE buffer (10 mM Tris HCl pH 8 and 1 mM EDTA). Furthermore, the DNA samples were stored at –20oC until being used for analysis.[Citation17]
Real Time PCR Amplification of Pork DNA Using D-Loop686 and cytb Primers
Amplification of pork DNA on variety of raw meat and dendeng was done using SsoFast®Evagreen®SUPERMIX with two sets of primer (). The reaction was performed at 30 cycle with the condition of denaturation at 95oC for 5 s, annealing at 62oC (D-Loop686 primer) and 61oC (cytb primer) for 10 s, and extention at 72oC for 10 s.[Citation12,Citation13]
TABLE 1 Sequence of primers at target region of mitochondrial D-Loop686 and Cytochrome b gene (cytb)
Test of Primer Specificity to Porcine DNA Using Real Time PCR
Gradient PCR was carried out using two sets of primers on raw pork meat at various annealing temperature (44.9–62oC). This work was done to find an optimum annealing temperatures.Test of specifity was conducted on five species of raw meats using optimum annealing temperature obtained. In addition, the specificity of primers on pork DNA from pork dendeng (positive control) was compared to that of DNA from beef dendeng (negative control) and DNA from mixed pork-beef dendeng.
Determination of Limit of Detection (LoD) of Porcine DNA
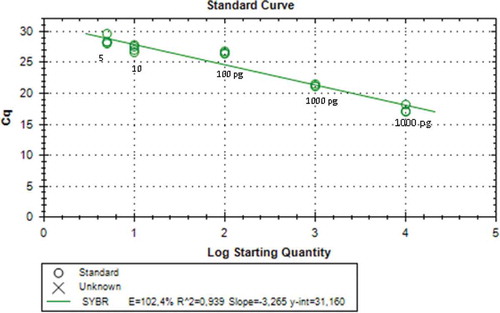
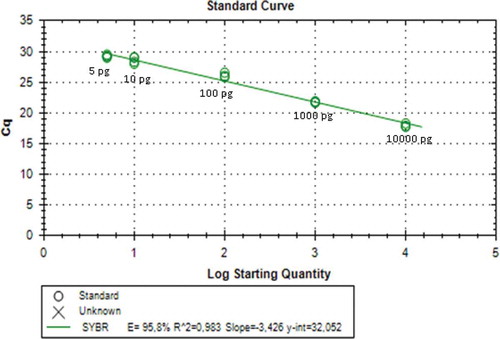
The LoD was determined by making standard curve of diluting series (10,000, 1000, 100, 10, 5, and 1 pg) of DNA sample of pork dendeng. Determination of LoD was also performed on the meat mixture of porcine-beef dendeng with concentration of 0.5, 1, 3, 5, and 10% w/w. The value of LoD was expressed as the amount of DNA amplified by primer resulting relative fluorescence unit of 100 measured using RT-PCR.
Detection of Pork Adulteration in Commercial Dendeng Products
Identification of pork adulteration in six commercial dendeng derived from traditional markets and supermarkets in Yogyakarta. The analysis was done using two sets of primers with the condition as indicated on amplification procedure.
RESULTS AND DISCUSSION
Species identification in meat products has attracted the attention of researchers in recent years. This is due to the rise of trade related to dubious food labeling. In this study, 114 bp of mitochondrial D-Loop and 149 bp of cytb fragments were amplified using D-Loop686 and cytb primers, respectively, to detect the presence of pork DNA in dendeng products.
Primer Specificity Analysis of Pork DNA Using Real Time PCR
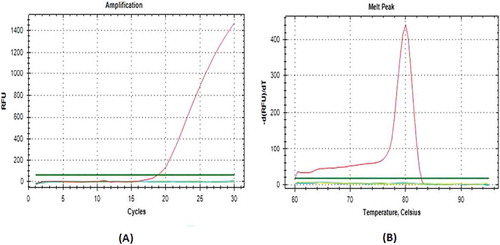
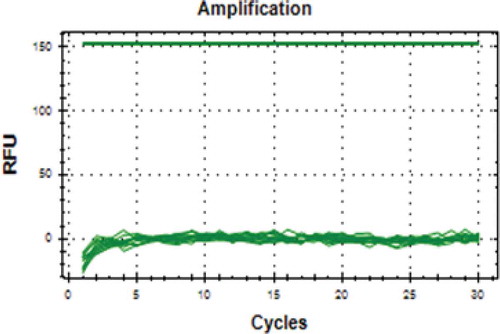
Annealing temperature is an important factor in the process of RT-PCR amplification, this is associated with a primary attachment of a primers on a target DNA, so that the amplification process can run specifically. Various different annealing temperature (44.9–59.9oC) was done for the D-Loop686 primers using thermal gradient RT-PCR (data not shown). However, it seemed that the primers are un-specifically bound on pork DNA, as indicated by the presence of two peaks on the melting curve profile, at 82 and 88oC (). This indicated that the primers amplified at other region, at chromosome 8, beside of the target sequence of mitochondrial D-Loop. Consistent result was also obtained when the annealing temperature was increased at 2–3 degrees above the melting temperature (Tm) of 59oC. At 62oC annealing temperature, however, the mitochondrial D-Loop686 showed high specificity to pork DNA, as indicated by no amplification results appeared on other species (beef, chicken, horse, and goat meat) at (). It was proven that this additional peak does not result from primer dimer, since no amplification product on sample without DNA template (data did not show). Therefore, the D-Loop686 primers designed by Himawati[Citation13] could be used to analyze pork adulteration found in various meat processing, including dendeng, since it only specifically amplified pork DNA. The primers of D–Loop686 and cytb genes have a high specificity, indicated by sequence similarity of the results of the basic local alignmet search tool (BLAST).
FIGURE 1 Amplification curves A: and Melting Peak; B: Specificity of pork Mitochondrial D-Loop686 primer on DNA from various raw meat. Red line: pork; yellow line: beef; dark blue line: chicken; green lines: goat; blue line: horse.
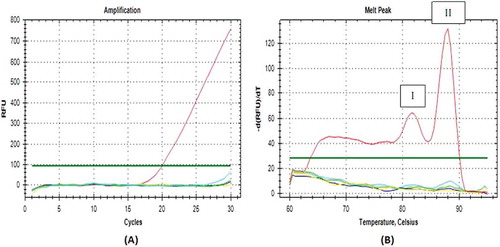
High specificity result was obtained when the amplification was performed using the cytb primers, designed, and validated by Soares et al.[Citation12] with the length of amplificated product of 149 bp at an annealing temperature of 61oC and melting point at 80oC (). Similar to the D-Loop686 amplification products, the primer of cytb gene only amplified the pork DNA, but not other meat species. This indicated that even the D-Loop686 primer bind to two different parts, it could be used for identification of pork adulteration on food products.
Determination of Porcine in Laboratory Prepared Dendeng Products
The implementation of D-Loop686 and cytb gene primers was carried out in order to identify pork DNA on laboratory prepared dendeng. The results indicated that mithocondrial D-Loop686 primer had high specificity on pork DNA target, as indicated by the ability of this primer to detect pork DNA until the concentration of pork 0.5% in beef dendeng product with the consistent curve profile (). The cytb gene primers also had similar ability with D-Loop686 primer to amplify pork DNA until the pork concentration of 0.5% in beef dendeng (). The results showed that these two primers, D-Loop region and the cytb gene had the capability to identify pork adulteration on the mixed pork-beef dendeng product at all variations of tested concentrations.
FIGURE 3 Amplification curves A: and Melt Peak Curve; B: Specificity of pork Mitochondrial D-loop686 primer on laboratory prepared dendeng. a: 100% Beef 100%; b: 100% Pork; c: 10% Pork-beef 10%; d: 5% pork-beef; e: 3% pork-beef; f: 1% pork-beef; g: 0.5% pork-beef.
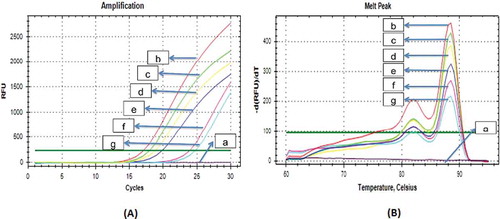
FIGURE 4 A: Curve Amplification and Melt Peak Curve; B: Specificity of pork ctyb on laboratory prepared dendeng. a: beef 100%; b: 100% pork; c: 10% pork-beef 10%; d: 5% pork-beef; e: 3% pork-beef; f: 1% pork-beef; g: 0.5% pork-beef.
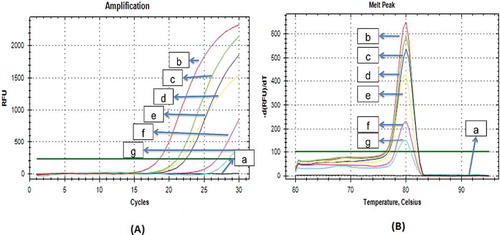
The specifity test of these two primers was also carried out on the pure pork dendeng as a positive control, and pure beef dendeng as a negative control. The result indicated that, mitochondrial D-Loop686 and cytb primers could amplify the pork DNA from pure pork dendeng, but do not amplify the beef DNA from pure beef dendeng.
Detection Limit and Repeatibility Assay
The standard curve showed that amplification in 100% pork dendeng using mitochondrial D-Loop686 and cytb primers had the lowest detection limit of 5 pg obtained from using of both primers. These results can be seen in and with coefficient determination (R2) value were of 0.983 and 0.939, respectively. The result of using D-Loop686 primer met the requirement of the determination coefficient that is 0.980,[Citation18] but it was not for cytb primer. The efficiency value (E) of D-Loop686 and cytb primers was 95.8 and 102.4%, respectively. Both values met the requirement of good reaction with a range of E-values from 90 to 110%.[Citation18] The standard curve showed that the amplification on the mixed dendeng of pork:beef with the concentrations of 0.5, 1, 3, 5, and 10% show that R2 value was 0.897 and E-value was 60.7% for using D-Loop686 primer, while R2 value of 0.764 and E-value of 66.4% for cytb primer were obtained. The R2 and E-values obtained did not meet the requirement as in Soares et al.[Citation12]
The repeatability test showed positive amplification from all DNA samples containing pork. The coefficient variation (CV) for real time PCR should be ≤25% of dynamic range of the method.[Citation19] A good CV was obtained with the value of 18.03% for D-Loop686 and 17.2% for cytb primer, respectively. Futhermore, the repeatability test was also performed on mixed pork-beef dendeng which resulted on CV value of 12.61% (for D-Loop686) and 10.56% (for cytb).
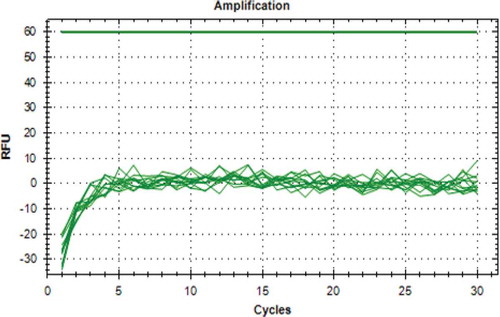
Detection of Pork Adulteration in Commercial Dendeng Products
The developed assay was further applied to the six commercial dendeng products for identification purposes. It was indicated that there was not amplification from all the samples. Therefore, all of the commercial dendeng products did not contain pork DNA as shown in and .
CONCLUSION
The primer of D-Loop686 region had high specificity to pork at fresh meat samples and mixture of pork:beef dendeng products with the lowest detection limit of 5 pg/µL from pork dendeng and 0.5% of mixture pork:beef dendeng as determined by the amount of DNA amplification relative to fluorescence unit of 100 measured using RT-PCR. A similar result was obtained from analysis of pork DNA using the cytb primer. The six commercial dendeng amplified using both primers showed no amplified products, meaning that these dendeng products do not contain pork.
FUNDING
This research was finnancially supported by a grant from the Director General of Higher Education, Ministry of Education and Culture, through project number LPPM-UGM/346/LIT/2014.
Additional information
Funding
REFERENCES
- Rahmawati, P. Analisis Cemaran daging Babi pada produk Dendeng Sai yang beredar di Wilayah Ciputat dengan metode Amplifikasi DNA menggunakan Real Time PCR [Analysis of Pork Contamination in Beef Dendeng Based on DNA Amplification using Real Time Polymerase Chain Reaction]. Skripsi, UIN Syarif Hidayatullah Jakarta, Indonesia, 2012.
- Rohman, A.; Che Man, Y.B. Analysis of Pig Derivatives for Halal Authentication Studies. Food Review International 2012, 28(1), 97–112.
- Che Man, Y.B.; Marina, A.M.; Rohman, A.; Al-Kahtani, H.A.; Norazura, D.A. FTIR Spectroscopy Method for Analysis of Palm Oil Adulterated with Lard in Pre-Fried French Fries. International Journal of Food Properties 2014, 17, 354–362.
- Che Man, Y.B.; Mat Hashim, D.; Mohamed, A.K.S. Rapid Detection Of Pork In Food Products By Electronic Nose For Halal Authentication. Proceedings on the 3rd IMT-GT International Symposium on Halal Science and Management, University Putra Malaysia (UPM), Malaysia, Dec 21–22, 2009.
- Rohman, A.; Sismindari Erwanto, Y.; Che Man, Y.B. Analysis of Pork Adulteration in Beef Meatball Using Fourier Transform Infrared (FTIR) Spectroscopy. Meat Science 2011, 88, 91–95.
- Kurniati, E.; Rohman, A.; Triyana, K. Analysis of Lard in Meatball Broth Using Fourier Transform Infrared Spectroscopy and Chemometrics. Meat Science 2014, 96, 94–98.
- Chen, F.C.; Hsieh, Y.H. Detection of Pork in Heat-Processed Meat Products by Monoclonal Antibody-Based ELISA. Journal Association of Official Agricultural Chemist International 2000, 83(1), 79–85.
- Che Man, Y.B.; Aida, A.A.; Raha, A.R.; Son, R. Identification of Pork Derivatives in Food Products by Species-Specific Polymerase Chain Reaction (PCR) for Halal Verification. Food Control 2007, 18(7), 885–889.
- Raharjo, T.J.; Cahyaningtyas, W.; Surajiman; I.; Pranowo, D. Validation of PCR-RFLP Testing Method to Detect Porcine Contamination in Chicken Nugget. Indonesian Journal of Chemistry 2012, 12(3), 302–307.
- Che Man, Y.B.; Mustafa, S.; Mokhtar, N.F.K.; Nordina, N.; Sazilia, A.Q. Porcine-Specific Polymerase Chain Reaction Assay Based on Mitochondrial D-Loop Gene for Identification of Pork in Raw Meat. International Journal of Food Properties 2012, 15(1), 134–144.
- Kesmen, Z.; Güllüce, A.; Yilmaz, M.T.; Yetiman, A.E.; Yetim, H. Taqman Based Duplex Real-Time Pcr Approach for the Detection And Quantification of Donkey and Pork Adulterations in Raw And Heat-Processed Meats. International Journal of Food Properties 2013. DOI:10.1080/10942912.2012.654569.
- Soares, S.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I. A SYBR Green Real-Time PCR Assay to Detect and Quantify Pork Meat in Processed Poultry Meat Products. Meat Science 2013, 94(1), 115–120.
- Himawati, A. Analisis Campuran Daging Babi dan Sapi dalam Bakso dengan Metode Spektrofotometri Fourier Transform Infrared dan Real Time PCR [Analysis of Pork-beef Mixture in Meatball using FTIR Spectroscopy and Real Time Polymerase Chain Reaction]. Master thesis, Universitas Gadjah Mada, Yogyakarta, Indonesia, 2013.
- Dooley, J.J.; Paine, K.E.; Garrett, S.D.; Brown, H.M. Detection of Meat Species Using Taq Man Real-Time PCR Assays. Meat Science 2004, 68(3), 431–438.
- Purnomo, H.; Buckle, K.A.; Edwards, R.A. Partial Substitution of Meat in Dendeng Giling with Breadfruit and Corn Grits. Journal of Food Science and Technology 1984, 21(5), 326–328.
- Meliany, H.P.; Budianta, T.D.W. Pemanfaatan Buah Pepaya Muda Dalam Pembuatan Dendeng Giling Kambing [The Use of Unriped Papaya Fruit in Preparation of Lamb Dendeng]. Jurnal Teknologi Pangan Dan Gizi 2001, 2(1), 7–12.
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning, 2; Cold Spring Harbor Laboratory Press: New York, NY, 1989.
- Biorad. Real Time PCR Application Guide from Bio Rad Free Ebooks (pdf, doc, xls files). http://ebookbrowsee.net/re/real-time-pcr-application-guide-from-bio-rad ( accessed September 2014).
- CAC/GL 74., Codex Guidelines on Performance Criteria and Validationof Methods for Detection, Identification, and Quantification of Spesific DNA Sequences and Spesific Proteins in Foods, 2010.