Abstract
This study was performed to compare the total protein and protease content among the whole fruit, flesh, and peel of nine different pear cultivars. Pear proteases were functionally characterized with respect to three enzyme assays. Proteases from pears were further identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, in gel activity staining, and matrix assisted laser desorption/ionization-time of flight mass spectrometry analysis. Flesh from Whasan, Nikita, and Hanareum cultivars contained relatively more total protein and protease and showed high enzyme activities, while Chuwhang contained the lowest amount of protein and protease activity. Protease content and enzyme activities found in the pear flesh or whole fruits were two to six times higher than those in the pear peel. Pear cultivars contained one or two protease bands with molecular weights of 36 kDa and/or 38k Da. The larger band was further identified as a cysteine proteinase with 70% homology to the pear cysteine protease from Pyrus pyrifolia.
INTRODUCTION
Pear fruit is one of the most widely consumed fruits in the world. One of the major species of pears, Asian pears (Pyrus pyrifolia) have been cultivated mainly in Eastern Asia including Korea, China, and Japan. As the pear is a seasonal fruit, it is typically eaten fresh and is often found in processed foods such as juice, purees, jellies, and jams. For many years, the pear has been used not only as one of the most common edible fruits but also as a meat tenderizer in food systems. Crude protein extract from Asian pears and its activity on meat tenderization have previously been reported.[Citation1–Citation3]
The proteolytic enzymes of plant origin extensively studied and specifically, plant cysteine proteases, are commonly used in the food industry for meat tenderization, bread manufacture, flavor improvement, and beer clarification.[Citation4] Plant cysteine proteases studied include papain (papaya), ficin (fig), bromelain (pineapple), zingibain (ginger), actinidin (kiwifruit), and cucumin (cucumis).[Citation5–Citation9] They are widely studied for their high proteolytic activities. But enzymes such as actinidin, bromelain, and papain showed practical limitations due to their non-uniform or over activity on meat proteins.[Citation10] Among them, pear protease was recently emphasized as the ideal meat tenderizer to prevent over decomposition or mushy spots in meat compared to bromelain or actinidin.[Citation10]
Previous studies reported that a pear cysteine protease was purified from Nikita (NK) and Chuwhang (CW) cultivars with a molecular weight between 30 kDa and 38 kDa.[Citation3,Citation11] Crude or partially purified pear protease was functionally characterized with respect to proteolytic properties on meat tenderization.[Citation1–Citation3,Citation10] The proteolytic activity of pear protein extract showed optimum pH at 5.3–7.0 and temperatures of 40–70°C on chicken actomysin.[Citation2,Citation11,Citation12] Recent studies reported that pear protein extract showed optimum proteolytic activity at 0.1% concentration in pork loins with improved meat tenderizing properties when used in combination with pineapple or kiwifruit extract.[Citation3,Citation10]
Negligible studies have been conducted to compare the protease activity and proteolytic function of major Asian pear cultivars (Pyrus pyrifolia). Previous studies investigated cysteine proteases from one or two pear cultivars and their proteolytic activity on chicken actomyosin.[Citation3,Citation11] In this article, for the first time, we compared pear proteases from nine major Asian pear cultivars and characterized their proteolytic functions with respect to three different enzyme assays.
MATERIALS AND METHODS
Pear Fruits Sampling and Extraction
Nine cultivars of Asian pear (Pyrus pyrifolia) were chosen for this study based on the highest production in Korea. These included CW, Shinwha (SW), Whasan (WS), Gamcheon (GC), Manpung (MP), Mansu (MS), Wonwhang (WW), NK, and Hanareum (HA) cultivars. All of the cultivars were picked from 15-year old trees that were located at an orchard of the Naju National Pear Experimental Station in the Chonnam province of Korea, and were harvested at the mature fruit stage only. Seven to 10 fruits from each of the selected standard pear trees were used for the experiments with uniformity in size and no defects. Protein from pear cultivars was extracted according to a previous study with slight modifications.[Citation13] Pear fruits were peeled and their cores were removed. The whole fruit, flesh, or peel were separately washed, rapidly cut into thin slices, and lyophilized by freeze-drying (FD8512, Ilshin, Korea). The samples were then ground by pulverizing (FM-681C, Hanil Electric., Korea) and stored at –20°C in polyethylene bags until analysis. Each pear cultivar powder (10 g) was mixed with extraction buffer (10 mM cysteine, 1 mM ethylenediamine tetraacetic acid (EDTA), 10 mM sodium phosphate, pH 6.5) and incubated by gently shaking at 4°C for 1 h. The extract was centrifuged at 4°C for 30 min at 15,000 × g (Avan-Tr J-E, Beckman Coulter, USA) to remove insoluble material. The supernatant was filtered through Whatman No. 1 filter paper and 0.45 μM membrane filter at 4°C and used as the pear protein extract.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) Analysis
Total protein in pear protein extracts was determined using a Pierce Chemical Co. protein determination kit based on a modified Bradford technique.[Citation14] Bovine serum albumin, in the range of 50–200 µg /mL, was used as the standard and the absorbance of samples at 595 nm was recorded using a Beckman-Coulter 7500 spectrophotometer. SDS-PAGE was carried out using a 5% stacking gel and 12% running gel under reducing and non-reducing conditions. Aliquots of pear protein extracts containing 20 μg of protein were mixed with the sample buffer (0.5 M Tris-HCl, pH 6.8, 10% SDS, 12% glycerol, 0.05% bromophenol blue) containing 2% β-mercaptoethanol and heated at 100°C for 5 min. Electrophoresis was performed at 4°C at 150 V using a mini-Protean II apparatus (Bio-Rad Laboratories). The molecular mass of protease in pear protein extract was estimated by comparison with the low range standard protein kit from Bio-Rad. The protein bands were visualized by staining with 0.1% Coomassie brilliant blue R-250. Protease in pear protein extracts were quantified by SDS-PAGE analysis and densitometry scanning of protein bands with molecular weights of 36 to 38 kDa in each lane using the National Institutes of Health (NIH) program (Scion Corporation). All samples were analyzed in triplicate.
Casein Gel Zymography and Protease Quantification
Proteolytic activities in pear protein extracts were detected by casein gel zymography.[Citation15] Samples for activity staining were prepared under native conditions without heat treatment or β-mercaptoethanol. Aliquots of extract containing 40 μg of protein were supplemented with electrophoresis non-reducing sample buffer. Samples were then separated under non-reducing conditions in a 12% polyacrylamide gel co-polymerized with 0.05% casein. Stacking gels contained 5% polyacrylamide. Electrophoresis was carried out with the same conditions described before for SDS-PAGE. Low range prestained standard proteins (Bio-Rad) were used as molecular weight markers. After electrophoresis, the gels were washed (3 × 30 min) in 2% Triton X-100, 0.05 M phosphate buffer pH 6.5 (washing buffer) in order to remove SDS, then incubated for 14 h at room temperature in developing buffer: 1% Triton X-100, 0.05 M phosphate buffer pH 6.5, 1 mM EDTA, 0.01 M cysteine. For the development of enzyme activity, the gels were stained with Coomassie brilliant blue R-250, destained in methanol–acetic acid–water solution. Proteolytic activities were detected as clear, unstained bands on a blue background.
Characterization of Esterase, Caseinolytic, and Collagenase Activities
Protein extracts from nine cultivars were assayed for esterase activity, caseinolytic activity, and collagenase activity. Esterase activity of the pear protein extract was investigated using the substrate CBZ-Lys-p-nitrophenyl (CBZ Lys-ONp) ester (Sigma, #A4341).[Citation9] Phosphate buffer, 50 mM pH 6.0 (450 μL) was first equilibrated for 3 min to room temperature in a microfuge tube. Then 10 μL of 20 mM CBZ-Lys-ONp ester in acetonitrile was added. Subsequently, each pear protein extract from nine cultivars (40 μL) was individually added into tubes while the same volume of distilled water was added to the control. Substrate hydrolysis was allowed to occur for 1 min. After incubation, 500 μL of 1 M sodium carbonate solution was added to each tube to quench the enzyme reaction. The samples were then centrifuged at 12,000 × g for 5 min and the supernatant was measured at 410 nm using a double beam ultraviolet (UV)-visible spectrophotometer (Biospec 1601; Shimadzu Scientific Instruments, Japan) in a 1 mL cuvette. One unit of enzyme activity (U) was defined as the amount of enzyme causing an increase in the absorbance by one unit at 410 nm under the above conditions.
Caseinolytic activity of the pear protein extract was determined using Quanti-protease Assay kit (Thermo Scientific #23263).[Citation16] The assay was based on the ability of proteases to hydrolyze succinlyated casein and the measurements were carried out using a microplate reader (Biotek Powerwave, USA). The 96-well plate was temperature equilibrated in an oven at 37°C for 3 min. A succinlyated casein solution (100 μL of 1 mg/mL) and pear protein extract (50 μL) were added. Then 50 mM sodium borate buffer, pH 8.5 (100 μL) and 2,4,6-trinitrobenzene sulfonic acid (TNBSA) solution (50 μL) were added to each well for purple color development over 20 min. The absorbance at 450 nm was read immediately after the plate was gently shaken for 3 s. U was defined as the amount of enzyme causing an increase in the absorbance by one unit at 450 nm under the above condition.
Collagenase activity of the pear protein extract was determined using collagen impregnated with azo-dye (Azocoll, Sigma, #A4341).[Citation17] Azo-dye labeled peptides released into the assay solution from the insoluble Azocoll matrix (in proportion to peptide bonds hydrolyzed) were measured at 520 nm. Azocoll (25 mg) was suspended in 12.5 mL of 50 mM phosphate buffer pH 6.0 and stirred for 2 h at room temperature, followed by decanting of the supernatant containing extracted azo-dye labeled collagen peptides that can interfere with the assay.[Citation17] The settled substrate was re-suspended in 12.5 mL of the same buffer and the washing step was repeated. Aliquots (1.0 mL) of the Azocoll suspension (in 12.5 mL) were stored at –20°C until use. Pear protein extract (100 μL) was added to 1.0 mL of Azocoll suspension and the tubes were tumbled at the incubation temperature at 37°C in a mini Hybaid oven. The samples were then centrifuged at 14,000 × g for 10 min and absorbances of the supernatants were measured at 520 nm after 24 h incubation. U was defined as the amount of enzyme causing an increase in the absorbance by one unit at 520 nm under the above conditions.
Protein Identification by In-Gel Digestion and Matrix Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF) Analysis
The sample preparation process followed the published method.[Citation18] Select bands in the SDS-PAGE gel were excised, cut into small pieces (1 mm3), and washed with 100 mL deionized water. The gel pieces were destained by adding 200 mL of a 2:1 (v/v) ratio of acetonitrile: 25 mM ammonium bicarbonate for 15 min, and this step was performed until the gel pieces were completely destained. The supernatant was removed and gels were then dehydrated by adding 200 mL acetonitrile for 15 min prior to drying in a vacuum centrifuge. Then 50 mL of a 10 mM dithiothreitol (DTT) solution in 100 mM ammonium bicarbonate was added, and the proteins were reduced for 1 hr at 56°C. After cooling to room temperature, the DTT solution was replaced with the same volume of 55 mM iodoacetamide in 100 mM ammonium bicarbonate and gels were incubated for 45 min at room temperature in the dark. The solution was then removed, the gel pieces were dehydrated in acetonitrile, and the solvent was evaporated before adding 10 mL of a trypsin solution (proteomics grade, Sigma; 10 ng/mL in 50 mM ammonium bicarbonate). After allowing the gel plug to swell for 15 min at 4°C, 30 mL of 50 mM ammonium bicarbonate was added and the digestion proceeded at 37°C overnight. The supernatant was then harvested following centrifugation at 10,000 × g for 1 min. The remaining peptides in the gel were extracted with a solution of 50% acetonitrile with 5% formic acid (v/v) for 10 min with shaking, and subsequently pooled with the supernatant and dried. The extracted tryptic peptides were subjected to matrix assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis. All collected MALDI-TOF MS data were processed and submitted to a MASCOT (http://www.matrixscience.com) search of the National Center for Biotechnology Information (NCBI) database (http://blast.ncbi.nlm.nih.gov). Proteins were identified through database search by using peptide mass fingerprints.
Statistical Analysis
All experiments were carried out in triplicate with three replicates and results were expressed as mean ± standard deviation. The data were analyzed by one-way analysis of variance (ANOVA), and the means of different groups were compared with the Duncan’s multiple range test (DMRT) using the SPSS version 17.0 statistical software package. Values of p < 0.05 were considered as significant in all cases.
RESULTS AND DISCUSSION
Total Protein Content of Nine Pear Cultivars
Total protein content in different segments of nine pear cultivars was determined and shown in . Crude protein from pear cultivars were extracted with 10 mM sodium phosphate buffer (pH 6.5) containing 10 mM cysteine, 1 mM EDTA followed by filtration.[Citation13] Cysteine (a reducing agent) and EDTA (a chelating agent) should be added to protein extracts since cysteine protease activities are extensively inhibited by metal ions and oxidation.[Citation19]
FIGURE 1 Total protein contents in a: whole fruit; b: flesh; and c: peel of nine pear cultivars in mg/g dry weight. Value are means ± SD (n = 3). Different letters (a–h) of each indicate significant difference within column at p < 0.05 by DMR test. The cultivars used were Chuwhang (CW), Shinwha (SW), Whasan (WS), Gamcheon (GC), Manpung (MP), Mansu (MS), Wonwhang (WW), Nikita (NK), and Hanareum (HA).
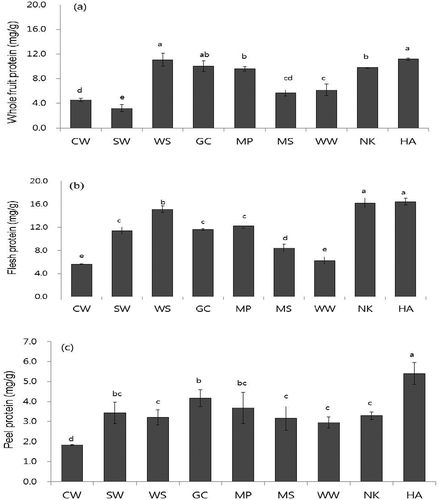
Nine cultivars of Asian pear (Pyrus pyrifolia) were chosen for this study since they are grown in Korea. These included CW, SW, WS, GC, MP, MS, WW, NK, and HA cultivars. The nine pear cultivars tested were found to have total protein contents ranging from 1.84 to 16.5 mg/g dry weight (DW; ). Total protein concentrations were highest in the flesh, followed by the whole fruit and peel, regardless of pear cultivars. Pear peel contained approximately 25–30% of the total protein compared to the flesh except WW.
Among the species tested, total protein contents were significantly higher in WS, NK, and HA cultivars but lower in CW and WW cultivars. In the flesh, WS, NK, and HA cultivars contained the highest protein content (15.16 ~ 16.42 mg/g DW), approximately three times greater than that of CW cultivar (). These three cultivars also showed higher protein contents in the whole fruit (9.78 ~ 11.17 mg/g DW) and peel (3.21 ~ 5.40 mg/g DW) among the cultivars ( and ). In the pear peel, GC pear also contained a higher protein content with 4.17 mg/g DW, similar to WS, NK, and HA cultivars ().
SDS-PAGE and Casein Gel Zymography Analysis
Protein profiles of flesh extracts of the nine pear cultivars were analyzed by SDS-PAGE analysis (). Crude protein extract (20 μg of protein) was denatured and reduced for SDS-PAGE but in native conditions for activity staining ( and ). Arrows indicate the putative protease bands of pear cultivars with the expected molecular weights of 36 ~ 38 kDa and one or two bands were visible with strong intensities in all the cultivars (). This pattern of cysteine proteases agrees with previous reports about the size of pear protease at 30 to 38 kDa,[Citation3,Citation11] and the pear proteases of the nine cultivars shown here were found as one or two bands (). Also, these results are in agreement with previous reports regarding multiple bands of cysteine protease with similar size in other fruits. Actinidin in kiwifruit existed in multiple isoforms with molecular weights of 24 ~ 30 kDa,[Citation20] and pineapple stem and fruit had at least five distinct cysteine proteases with molecular weights of 26 ~ 28 kDa.[Citation21,Citation22] Fig latex also had multiple ficins with molecular weights of 23 ~ 25 kDa.[Citation23] Among pear cultivars, five cultivars including SW, WS, GC, NK, and HA exhibited two bands with expected molecular weights of 38 and 36 kDa while other cultivars only showed one band with a molecular weight of 38 kDa. The band intensities were significantly high in WS, NK, and HA but lower at CW ().
FIGURE 2 SDS-PAGE; a: analysis; and b: native-PAGE zymography of nine pear cultivars. M: molecular weight marker. Arrow indicates the protease bands of pear cultivars with the expected molecular weights of 36 and 38 kDa. The cultivars used were Chuwhang (CW), Shinwha (SW), Whasan (WS), Gamcheon (GC), Manpung (MP), Mansu (MS), Wonwhang (WW), Nikita (NK), and Hanareum (HA).
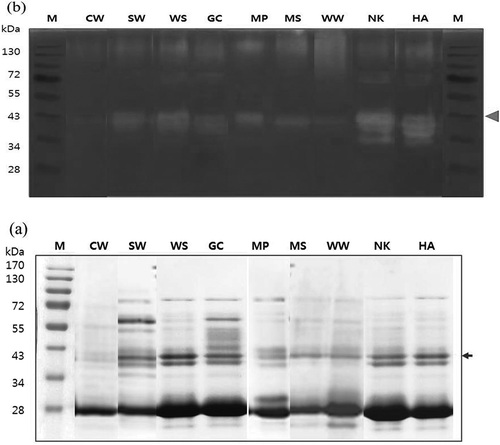
To verify the band of proteolytic enzymes, activity staining was performed using substrate (casein) gel electrophoresis. Since SDS-casein-PAGE is an effective method to identify specific proteolytic activities, protease activities of nine pear cultivars were characterized by zymogram analysis after SDS-casein-PAGE (). The clear zones of proteolytic activity on the dark background appeared at protein bands of 36 to 38 kDa (). WS, NK, HA, SW, and GC cultivars showed both bands, while MP, MS, WW, and CW cultivars showed only one band (). This observation was positively correlated with the protease patterns in SDS-PAGE analysis of pear cultivars (). The protease bands in casein gel zymography appeared with slightly higher molecular weights because the casein substrate in the gel might interfere with the electrophoretic migration and the proteins were not reduced.
With respect to the number of pear protease, several factors can be considered such as auto digestion or the properties of pear cultivars. There is the possibility for pear protease to undergo auto digestion because proteolytic enzymes like ficin or bromelain use native proteolytic proteins as a substrate for hydrolysis.[Citation23] Pears could have one or two proteases, depending on the cultivar. Previous work reported that the number and relative amounts of the ficins were differed among the cultivars or varieties of fig latex (F. carica).[Citation23,Citation24]
Characterization of Esterase, Caseinolytic, and Collagenase Activities
To achieve a more complete vision of the protease activity of the whole fruit, flesh, and peel extracts, three different assays were used in this study: esterase, caseinolytic activity, and collagenase. The results are shown in –, and significant differences (p < 0.05) were observed among the nine pear cultivars detected. Esterase assays are often used to provide an initial indication of the catalytic activity of cysteine proteases such as actinidin, papain, or ficin.[Citation16,Citation25] In this study, the ester hydrolysis activities of the protein extracts from nine pear cultivars were investigated with the substrate CBZ-Lys-ONp (). The average values and standard deviations of esterase activity of the pear cultivars are presented in . Some variations (p < 0.05) in esterase activity were noted among the nine pear extracts, with some exceptions.
TABLE 1 Esterase activity of nine pear cultivars
TABLE 2 Caseinolytic activity of nine pear cultivars
TABLE 3 Collagenase activity of nine pear cultivars
The pear cultivars were found to have esterase activity ranging from 172.2 to 25.2 U/mg protein (). Esterase activity of pear protein extract showed a similar pattern to that of protein contents with higher values of 172.2 to 58.3 U/mg protein in pear flesh, but lower values of 50.6 to 25.2 U/mg protein in pear peel ( and ). In general, the determined values of protease activity found in flesh were remarkably higher than those in peels by approximately three to four times. This agrees with the research reporting concentrations of protease in the flesh of the oriental pear, kiwifruit, and mango that were higher than in the peel.[Citation9,Citation10,Citation26]
Esterase activity of pear protein extract among cultivars showed a similar trend to that of casein gel zymography analysis. For the flesh, esterase activities ranged from 58.3 to 172.2 U/mg protein, with the highest in HA pear, and the lowest in CW pear. For the whole fruit and peel, activities of the nine pear cultivars have similar trends to those of flesh. Especially, the cultivars WS, NK, HA, and GC showed high amounts of esterase activity. It is worth mentioning that GC peel contained statistically higher esterase activity than the other pears with the value of 50.6 U/mg protein. CW pear showed the lowest protease activity and protein content of all the pears (). Overall, esterase activities in protein extracts were different by two to three times among the pear cultivars. Although ester compounds have been used extensively in protease activity analysis,[Citation16,Citation27] these compounds do not necessarily represent the true kinetic behavior of an enzymes’ ability to hydrolyze amide bonds in protein polymers. Hence, succinlyated casein was employed to assess the proteolytic activity (amide bond hydrolysis) of the pear protein extracts.
The pear cultivars were found to have caseinolytic activity ranging from 10.8 to 96.9 U/mg protein (). Caseinolytic activity of pear protein extract showed a similar pattern to that of esterase activity with pear flesh containing higher values of 14.9 to 96.9 U/mg protein and peel with lower values of 10.8 to 27.3 U/mg protein. Among the cultivars, WS, NK, and HA flesh showed statistically higher caseinolytic activity of 80.0 ~ 96.9 U/mg protein and CW flesh showed lower caseinolytic activity of 14.9 U/mg protein. Among the cultivars, pear protein extracts of whole fruit and peel showed a similar proteolytic activity pattern with those of flesh except for GC pear. The GC peel contained the highest caseinolytic activity with a value of 17.3 U/mg protein. Here, the CW pear also showed very low activity which was 16, 18, or 40% of the activity of NK pear in whole fruit, flesh, or peel, respectively. Overall, caseinolytic activities in protein extracts were different by 2.5–6 times among the cultivars.
According to previous work, proteolytic activity of pear protein extract with casein substrate (20 μM or 40 μg/mL) was similar to that from papaya or fig but 25 ~ 30% lower than that of kiwifruit or pineapple.[Citation10,Citation12] In this study, proteolytic activities of nine pear extracts (80 ~ 96 μg/mL) is more than twofold higher than that found in other studies,[Citation10,Citation12] indicating that our pear extract has a higher specificity for casein.
Collagen is a right handed supercoil and contains three polypeptide (α) chains held together by inter-chains hydrogen bonds. Collagen hydrolysis capability of cysteine proteases such as bromelain or actinidin have been used to obtain kinetic parameters.[Citation16,Citation28] Collagenase activity of the pear extract was tested with azocoll, a commercially available azo dye-labeled collagen, which is a commonly used substrate for collagenase assays.[Citation29] According to a previous study,[Citation17] azocoll stock suspensions were extensively pre-washed to reduce non-specific release of azo dye. Azocoll collagen solution was kept suspended by agitation during the assay time course since agitation has been shown to have a critical effect on Azocoll hydrolysis.
Azo-collagen was employed to monitor the collagenase activity of the pear protein extracts from nine cultivars (). Collagenase activities of the nine pear cultivars ranged from 0.87 to 6.06 U/mg protein (). Collagenase activity pattern agreed with esterase or caseinolytic activity in whole fruit and flesh but differed from that in the peel. Whole fruit showed higher values with 1.67 to 6.06 U/mg protein than those of flesh or peel with 2.80 to 4.88 U/mg protein or 0.87 to 1.75 U/mg protein, respectively.
For the whole fruit and flesh, collagenase activity was higher in WS, NK, and HA cultivars (4.63 to 6.06 U/mg protein), approximately 3.5 times greater than in the CW pear. Enzyme activities in the peel of GC and NK pears were relatively high among cultivars but that in CW peel was very low. Interestingly, the HA peel showed lower collagenase activity with 1.05 U/mg protein, differing from flesh or whole fruit with high collagenase activity. Collagenase activities in pear extracts were different by 1.7–3.5-folds among the cultivars ().
With respect to collagenase activity, the values obtained () were comparable with the azo-collagen hydrolytic activities in other cysteine protease, such as actinidin (5 mg), papain (2 mg), bromelain (70 mg), and zingibain (3 mg).[Citation16] The collagenase activity in the nine cultivars (0.87 to 6.06 mg) was similar with other proteases but tenfold lower than that of bromelain.[Citation16,Citation25]
Cysteine Protease Identification
Pear protease was identified by in-gel trypsinization and MALDI-TOF analysis (). Conventional SDS-PAGE coupled with mass spectrometric analysis has been frequently used in proteomics protocols to identify proteins.[Citation30] Protein extract from NK pear was analyzed by SDS-PAGE and stained with Coomassie brilliant blue R-250 (). The arrow indicates the two bands of pear proteases excised with molecular weights of 36 and 38 kDa used for identification.
FIGURE 3 Identification of pear protease by in-gel trypsinization and MALDI-TOF analysis. a: Coomassie-stained gel of Nikita pear. Arrow indicates the bands excised for in gel digestion. b: Peptide mapping spectrum. Numbered peaks are matched mass peaks with expected sequence of cysteine protease. c: Peptide mass peak list. d: Sequence coverage through the peptide finger printing analysis by MALDI-TOF.
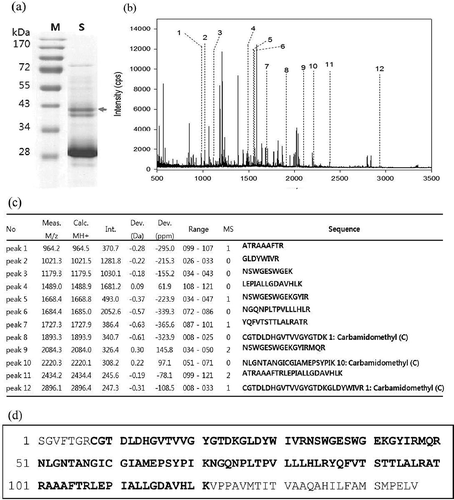
The peptide mapping spectrum of the upper protein band obtained from the gel is shown in . A total of 12 mass peaks contained matched peptides with the sequence of pear cysteine protease and peptide data of the 12 peaks (upper band) are presented in . Sequence coverage data through the peptide finger printing analysis by MALDI-TOF is summarized in . Amino acid sequence of resultant peptides from the upper band showed a 70% sequence homology to a cysteine proteinase from the Asian pear Pyrus pyrifolia in MASCOT searches of the NCBI database. Among the two bands with protease activity, only the upper band was successfully identified as a cysteine protease and the lower band exhibited no significant homology with any other reported plant peptides (data not shown). It could be reasoned that lower protein band may not be a protease or there is not enough sequence data of cysteine proteases in the NCBI database.
Cysteine Protease Contents of Nine Pear Cultivars
Cysteine proteases content in different segments of nine pear cultivars were determined (). Protease in total protein was quantified by SDS-PAGE analysis and densitometry scanning of selected protein bands with molecular weights of 36 and 38 kDa. The nine pear cultivars tested were found to have cysteine protease contents ranging from 0.07 to 4.76 mg/g DW (). The protease contents followed a similar trend to that of total protein, with higher values of 4.76 to 0.68 mg/g DW in pear flesh, but lower values of 0.07 to 0.83 mg/g DW in pear peel. This agrees with previous research reporting higher protease content in pear flesh and kiwifruits compared to the whole fruit or peel.[Citation3,Citation9,Citation11]
TABLE 4 Cysteine protease contents of nine pear cultivars
Among the species tested, cysteine protease contents were significantly higher in the WS, NK, and HA cultivars but lower in the CW and MS cultivars. In flesh, the WS, NK, and HA cultivars contained the highest protease content (3.67 ~ 4.76 mg/g DW), approximately two times greater than in the GC or SW cultivars and six times greater than in the CW or WW cultivars (). Those three cultivars also showed higher proteases content in the whole fruit (1.65 ~ 2.36 mg/g DW) and peel (0.35 ~ 0.58 mg/g DW) among the pear cultivars ().
NK, the representative cultivar with above 70% domestic production, had a higher value for cysteine protease in flesh and peel. Similar with total protein (), the GC cultivar showed higher content (0.83 mg/g DW) in the peel, but lower content in the whole fruit or flesh compared to the other cultivars. This is because GC has a relatively heavier weight than other cultivars, at more than 800 g, resulting in higher levels of protease in the peel due to the larger fruit volume.
With respect to cysteine protease, the values obtained () were comparable with the protease contents in other fruit, such as actinidin from kiwifruit (50% of total protein), papain from papaya latex (7 ~ 9% of total protein), and ficin from fig latex (9% of total protein).[Citation19,Citation23,Citation31] The protease contents in the nine cultivar pears (12 ~ 28% of total protein) were half the amount of that from kiwifruit but three times higher than that from papaya latex or fig latex reported.[Citation19,Citation23,Citation31]
CONCLUSIONS
The present study is the first evaluation of the protease in the whole fruit, flesh, and peel of nine different pear cultivars. Pear proteases from nine cultivars were characterized by three different activities including esterase, protease and collagenase. Cysteine proteases from pears were further identified by SDS-PAGE, in-gel activity staining, and MALDI-TOF analysis. Flesh from the WS, NK, and HA cultivars contained significantly more total protein and protease and showed higher enzyme activities, while CW appeared to have the lowest protease content. Protease contents and enzyme activities found in the pear flesh or whole fruits were approximately three to six times higher than those in the pear peel. Results of SDS-PAGE and in-gel activity staining analysis showed that pear cultivars contained one or two protease bands with molecular weights of 36 kDa and/or 38kDa. Peptide mass fingerprint using by MALDI-TOF of protease bands revealed that upper band matched a cysteine proteinase (Pyrus pyrifolia) with 70% homology but there was no identification of the lower band. These results indicate that WS, NK, and HA cultivars could be useful for the application of meat tenderization since they have high proteolytic activity.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Park Ji-Won from the Emass Corporation Institute for protease identification assistance.
FUNDING
This work was carried out with the financial support of the Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ01002403) at the Rural Development Administration, Republic of Korea.
Additional information
Funding
REFERENCES
- Choe, I.S.; Park, Y.J. A Study on Utilization As Meat Tenderizer from Korean Pear Protease. Korean Journal for Food Science of Animal Resources 1996, 16, 89–93.
- Choe, I.S.; Park, Y.J.; Ishioroshi, M.; Samejima, K. A New Protease in Korean Pears As Meat Tenderizer. Animal Science Technology 1996, 67, 43–46.
- Han, S.K.; Chin, K.B. Study on Meat Tenderness of a Protease Extracted from Domestic Pear. Korean Journal for Food Science of Animal Resources 2004, 24, 326–328.
- Kumar, C.G.; Takagi, H. Microbial Alkaline Proteases: From a Bio-Industrial Viewpoint. Biotechnol Advances 1999, 17, 561–594.
- Chaiwut, P.; Pintathong, P.; Rawdkuen, S. Extraction and Three-Phase Partitioning Behavior of Proteases from Papaya Peels. Process Biochemistry 2010, 45, 1172–1175.
- Kee, H.J.; Hwang, Y.S.; Kim, J.H.; Hog, Y.H. Application Fig Protease to Foods. Korean Journal for Food Science of Animal Resources 1998, 18, 19–26.
- Lee, D.H.; Jin, B.H. Encapsulation of Bromelain in Liposome. Journal of food Science and Nutrition 2001, 5, 81–85.
- Karnchanatat, A.; Tiengburanatam, N.; Boonmee, A.; Puthong, S.; Sangvanich, P. Zingipain, a Cysteine Protease from Zingiber Ottensii Valetin Rhizomes with Antiproliferative Activities Against Fungi and Human Malignant Cell Lines. Preparative Biochemistry and Biotechnology 2011, 41, 138–153.
- Nam, S.H.; Walsh, M.K.; Yang, K.Y. The Enzymatic Properties of Actinidin from Kiwifruit. Food Science and Biotechnology 2006, 15, 453–457.
- Kim, E.M.; Choe, I.S.; Hwang, S.G. Effects of Singular Manner or Mixed Type Treatment of Proteases Isolated from Pear, Pineapple and Kiwifruit on Actomyosin Degradation. Korean Journal for Food Science of Animal Resources 2003, 23, 193–199.
- Li, Q.H.; Mandai, P.K.; Lim, H.K.; Baatartsogt, Q.; Lee, C.H.; Jeon, G.J.; Choe, I.S.; Choi, K.D. Purification and Characterization of a Protease from Korean Pear (Pyrus Serotine L.) As Meat Tenderizer. Korean Journal for Food Science of Animal Resources 2009, 29, 157–163.
- Bai, Y.H.; Roh, J.H. The Properties of Proteolytic Enzymes in Fruits (Pear, Kiwifruit, Fig, Pineapple, and Papaya). Korean Journal of Food and Cookery Science 2000, 16, 363–366.
- Soares, P.A.G.; Vaz, A.F.M.; Correia, M.T.S.; Pessoa, A.; Carneiro-da-Cunha, M. Purification of Bromelain from Pineapple Wastes by Ethanol Precipitation. Separation and Purification Technology 2012, 98, 389–395.
- Bradford, M.M. A Rapid and Sensitive Method for Quantification of Microgram Quantities of Proteins Utilizing the Principle of Protein Dye-Binding. Analytical Biochenistry 1976, 72, 248–254.
- García-Carreño, F.L.; Dimes, L.E.; Haard, N. Substrate-Gel Electrophoresis for Composition and Molecular Weight of Proteinases of Proteinaceous Proteinase Inhibitor. Analytical. Biochemistry 1993, 214, 65–69.
- Ha, M.; Bekhit, A.D.; Carne, A.; Hopkins, D.L. Characterization of Commercial Papain, Bromelain, Actinidin, and Zingibain Protease Preparations and Their Activities Toward Meat Proteins. Food Chemistry 2012, 134, 95–105.
- Ha, M.; El-Din Bekhit, A.; Carnea, A.; Hopkins, D. Characterisation of Kiwifruit and Asparagus Enzyme Extracts, and Their Activities Toward Meat Proteins. Food Chemistry 2013, 136, 989–998.
- Kang, S.U.; Lubec, G. Complete Sequencing of GABAA Receptor Subunit B3 by a Rapid Technique Following in-Gel Digestion of the Proteins. Electrophoresis 2009, 30, 2159–2167.
- Chaiwut, P.; Nitsawang, S.; Shank, L.; Kanasawud, P. A Comparative Study on Properties and Proteolytic Components of Papaya Peel and Latex Proteases. Chiang Mai Journal of Science 2007, 34, 109–118.
- Pastorello, E.A.; Conti, A.; Pravettoni, V.; Farioli, L.; Rivolta, F.; Ansaloni, R.; Ispano, M.; Incorvaia, C.; Giuffrida, G.G.; Ortolani, C. Identification of Actinidin As the Major Allergen of Kiwi Fruit. Journal of Allergy and Clinical Immunology 1998, 101, 531–537.
- Napper, A.D.; Bennett, S.P.; Borowski, M.; Holdridge, M.B.; Leonard, M.J.; Rogers, E.E.; Duan, Y.; Laursen, R.A.; Reinhold, B.; Shames, S.L. Purification and Characterization of Multiple Forms of the Pineapple Stem-Derived Cysteine Proteinases Ananain and Comosain. Biochemical Journal 1994, 301, 727–735.
- Rowan, A.D.; Buttlet, D.J.; Barrett, A.J. The Cysteine Proteinases of the Pineapple Plant. Biochemical Journal 1990, 266, 869–875.
- Zare, H.; Moosavi-Movahedi, A.A.; Salami, M.; Mirzaei, M.; Saboury, A.A; Sheibanid, N. Purification and Autolysis of the Ficin Isoforms from Fig (Ficus Carica cv. Sabz) Latex. Phytochemistry 2013, 87, 16–22.
- Devaraj, K.B.; Kumar, P.R.; Prakash, V. Purification, Characterization, and Solvent-Induced Thermal Stabilization of Ficin from Ficus Carica. Journal of Agricultural and Food Chemistry 2008, 56(23), 11417–11423.
- Chalabi, M.; Khademi, F.; Yarani, R.; Mostafaie, A. Proteolytic Activities of Kiwifruit Actinidin (Actinidia Deliciosa cv. Hayward) on Different Fibrous and Globular Proteins: A Comparative Study of Actinidin with Papain. Applied Biochemistry and Biotechnology 2014, 172, 4025–4037.
- Mehrnoush, A.; Norashikin, A.; Chuan, L.T.; Ping, T.C; Hamed, M. Optimisation of Serine Protease Extraction from Mango Peel (Mangifera Indica cv. Chokanan). Food Chemistry 2011, 124, 666–671.
- Pickersgill, R.W.; Sumner, I.G.; Collins, M.E.; Goodenough, P.W. Structural and Electrostatic Differences Between Actinidin and Papain Account for Differences in Activity. Biochem J 1989, 257(1), 310–312.
- Lima, C.A.; Freitas Jr., A.C.V.; Lima Filho, J.L.; Converti, A.; Vuana Marques, D.A.; Carneiro-da-Cunha, M.G.; Porto, A.L. Two-Phase Partitioning and Partial Characterization of a Collagenase from Penicillium Aurantiogriseum URM4622: Aplication to Collagen Hydrolysis. Biochemical Engineering Journal 2013, 75, 64–71.
- Rowe, H.A.; Brown, M. Practical Enzyme Kinetics: A Biochemical Laboratory Experiment. Journal of Chemical Education 1988, 65, 548–549.
- Chung, W.G.; Cristobal, L.; Claudia, M.; Maier, S. Detection of Carbonyl-Modified Proteins in Interfibrillar Rat Mitochondria Using N0-Aminooxymethylcarbonylhydrazino-Dbiotin As An Aldehyde/Keto-Reactive Probe in Combination with Western Blot Analysis and Tandem Mass Spectrometry. Electrophoresis 2008, 29, 1317–1324.
- Arshad, Z.; Amid, A.; Yuso, F.; Jaswir, I.; Ahmad, K.; Loke, P. Bromelain: An Overview of Industrial Application and Purification Strategies. Applied Microbiology and Biotechnology 2014, 98, 7283–7297.
