Abstract
Adulteration of meat in processed food is a sensitive issue since certain meat species are prohibited in some religions such as Islam and Judaism. Some meat types are also potential carrier of some deadly diseases such as severe acute respiratory syndrome, anthrax, and hepatitis. Furthermore, unconscious consumption of certain meat might lead to an allergic reaction. Feline meat is not only taboo in most cultures but also under religious prohibition. However, cat meat has been consumed in certain countries including Cambodia, South Korea, China, and Vietnam. Several polymerase chain reaction assays are proposed for the detection of feline meat. However, those assays are not tested under processed food matrices. They are also based on longer targets (672, 331, 274, 180, and 108 bp) which breakdown under compromised states. Here we documented a very short-amplicon-length (69 bp) polymerase chain reaction assay and produced strong evidence that short targets are more stable than the longer ones. Feline specificity was confirmed by cross-challenging against 17 non-target species and target stability was tested after boiling, microwaving, and autoclaving treatments under complex matrices. The tested detection limit was 0.01% (w/w) of feline meat in ternary mixtures and 0.1% (w/w) in cooked meatballs.
Introduction
The huge turnover (2.3 trillion US dollars in 2012) in global food markets reflects that halal food consumption has rapidly expanded over the years.[Citation1] Ready-made foods, such as burgers, pizzas, hot dogs, sandwiches, meatballs, soups, cookies, candies, creams, and numerous others, are getting enormous popularity among the working and teenage population because of their convenient features and availability in roadside restaurants and groceries.[Citation2] However, recent horse meat scandals in Europe and[Citation3] rat and pig meat scandals in China,[Citation1] have affected adversely on the consumers trust on the labeled foods and their ingredients. Actually, the everyday happenings around us have made us feeling increasingly unsecured to protect our religious faiths, health, money, and wildlife. The consumption of feline meats or materials is “taboo” in many countries and also prohibited in certain religions such as Islam and Judaism. They are also potential carriers of hepatitis, severe acute respiratory syndrome (SARS), anthrax, and some other deadly zoonoses.[Citation4] However, cat meat has been consumed for years in several countries such as Cambodia, China, Thailand, South Korea, and Vietnam.[Citation5] Cat meats were sold as rabbit meats in eastern China[Citation6] and was served as Indian curry in the United Kingdom.[Citation7] Since the cat population is huge in most parts of the world and its meat does not have any value in legal markets, cat meat could be considered as a highly potential adulterant in halal, kosher, and other foods. Minced meats are frequently used in food products such as meatballs, burgers, and frankfurters,[Citation8] and targeted for adulteration for long time.[Citation9] Meatballs, which are made up with comminuted meats, are very popular across the world including Malaysia, China, Vietnam, Indonesia, the United States, India, and certain regions of Europe.[Citation8,Citation10] It could be formulated with pork, chicken, beef, and fish muscles.[Citation11] To survive in competitive markets and extra profit, lower valued meats such as pig and horse meats were replaced in frozen chicken meatballs,[Citation12] while chicken and turkey were substituted in pure beef meatballs.[Citation11] However, being a less discussed issue, feline meat adulteration in meatball formulations has not been independently verified and well reported. Several polymerase chain reaction (PCR) assays have been proposed for feline species detection.[Citation13–Citation18] Those assays were based on longer-length DNA fragment target (≥180 bp) which may break down under common food processing condition.[Citation19] Therefore, a convincing scientific evidence to prove this hypothesis has been missing in literature for a long time. Further, published assays have not been optimized under complex matrices of commercially processed foods. For the first time, we developed and optimized a 69-bp amplicon-length PCR assay for the detection of feline meats under complex and commercial matrices with evidence that it is superior to the published reports for detecting feline materials even from the highly processed foods.
Materials
Sample Collection
Fresh meat samples of beef (Bos taurus), water buffalo (Bubalus bubalis), chicken (Gallus gallus), domestic duck (Anas platyrhynchos), turkey (Meleagris gallopavo), sheep (Ovis aries), goat (Capra hircus), rat (Rattus norvegicus), pigeon (Columba livia), pig (Sus scrofa), cuttle (Sepia officinalis), carp (Cyprinus carpio), tilapia (Oreochromis aureus), shrimp (Gadus morhua), onion (Allium cepa), tomato (Solanum lycopersicum), and wheat (Triticum aestivum) were collected in triplicates on three different days from various wet markets and super markets across Malaysia (). Cat (Felis catus) meat samples from three different animals were collected from Jabatan Kesihatan Dewan Bandaraya Kuala Lumpur (DBKL), Air Panas Kuala Lumpur and Faculty of Veterinary Sciences in University of Putra Malaysia in Selangor. The species authenticity of the animal and plant species were verified by a taxonomy expert and transported to the laboratory under ice-chilled environment (4°C) and stored at (–20°C) until further use.
TABLE 1 History and sources of species and samples analyzed
Preparation of Ternary Admixtures and Meatballs
In order to apply the developed feline specific PCR assay for meat mixture analysis, ternary admixes (cat, beef, and wheat flour) was prepared according to Ali et al.[Citation2] Briefly, 10, 5, 0.2, and 0.01% (w/w) cat meat spiked ternary admixtures were made by adding cat meat in the ratio of (cat: beef: wheat flour) 10:45:45, 5:49.5:49.5, 0.2:49.9:49.9, and 0.01:49.995:49.995, in 100 g specimens. Homogenous semi-solid slurry was shaped by adding 100 mL distilled water and grinding properly in a food processing blender. On the other hand, pure meatballs were prepared according to Rohman et al.,[Citation10] by mixing a proper amount of ground beef, chicken, and cat meat with tapioca starch, cooking salt, garlic, and other spices as described in . For adulteration analysis, 100 g of beef and chicken meat were mixed with specified amount (10, 5, 1, 0.1, and 0.01% (w/w) of cat meat to formulate cat meat contaminated meatballs in the ratio of 10:45:45, 5:47.5:47.5, 1:49.5;49.5, 0.1:49.95:49.95, and 0.01:49.995:49.995, respectively. Finally, all the ingredients were blended properly and divided into four parts and each part was given mechanically into a ball shape.[Citation8] To mimic standard boiling and extensive autoclaving effect, all the admixtures and prepared meatballs were subjected to boiling at 100°C for 90 min and autoclaved at 120°C under 45-psi pressure for 2.5 h. All the admixtures and meatball samples were prepared on three different days by three independent analysts and DNA obtained from all samples were kept under –20°C for PCR analysis.
TABLE 2 A list of components commonly used to prepare beef, chicken, and cat meatballs (100 g) in this experiment
TABLE 3 Pairwise distances between 69 bp cytb feline-specific site and common meat, fish, and plant species potentially found in meatball formulations
Processing Treatment
Different types of processing and cooking treatments were applied to the collected meat samples to verify the stability of the target region. To simulate normal cooking, the fresh meat of target species was cut into small pieces (3 g) and then the meat was cooked under three different microwave cooking stages namely low (300 W), medium (500 W), high (700 W) microwaving for 30 min and boiling at 100°C for 90 min.[Citation19] Additionally, to comply with the European legislation,[Citation20] meat was autoclaved at 120°C for 50 min under 14.5 psi, 110°C for 2 h under 14.5 psi, and 133°C for 20 min under 43.51 psi. Consequently, extensive autoclaving was completed at 120°C for 2.5 h under 45 psi,[Citation19] to check the target gene stability under extreme processing treatments.
DNA Extraction
Total genomic DNA was extracted from 35 mg of raw, admixed, and treated samples of each species using Yeastern Genomic DNA Mini Kit (Yeastern Biotech Co., Ltd., Taipei, Taiwan) following manufacturer’s instructions. DNA from plant sources like onion, tomato, and wheat and commercial samples such as beef and chicken meatballs was extracted using cetyl trimethyl ammonium bromide (CTAB) method according to the Ma et al.[Citation21] Measurement of DNA concentration and purity was checked by NanoPhotometer® (IMPLEN, Nano Life Quest Sdn. Bhd; Selangor, Malaysia) taking absorbance at 260–280 nm and measuring A260/280 ratios. The extracted DNA was kept at –20°C until further use.
Primer Design
Feline-specific primers (Forward-5′-ACTATTATTTACAGTCATAGCCACAGC-3′) and (Reverse-5′-CAGAAGGACATTTGGCCTCA-3′) were developed targeting a 69 bp site of mitochondrial cytb gene using widely accessible Primer3plus software (www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) following Ali et al.[Citation19] Briefly, the selected primers were screened for unique feline specificity in an in silico analysis using online basic local alignment search tool (BLAST) against non-redundant nucleic acid sequences in National Center for Biotechnology Information (NCBI) data base (http://blast.ncbi. nlm.nih.gov/Blast.cgi). The expected target sequence was multiple aligned with five common halal animal meat species like beef, water buffalo, chicken, goat, and sheep; three avian species namely domestic duck, pigeon, and turkey; four fish samples including carp, cuttle, tilapia, and shrimp; three plant origin known as onion, tomato, and wheat; and two non-halal meat species such as rat and pork by ClustalW sequence alignment program (http://www.genome.jp/tools/clustalw/) to identify the variability of the primer binding regions and total mismatch between target and non-target species. The successfully designated primers were synthesized and supplied by the 1st BASE Laboratories Sdn Bhd (Selangor, Malaysia).
PCR Assay Optimization
The PCR reaction was performed in a gradient thermal cycler (Veriti, Applied Biosystems, USA) with 25 μL of reaction mixture containing of 5 µL of 5x Green GoTaq Flexi Buffer (Promega, Corporation, Madison, USA), 1.5 µL of 25mM of MgCl2 (Promega), 0.5µL of 0.2 mM of each dNTPs mix (sodium salts of dATP, dCTP, dGTP, and dTTP in water; Promega, Madison, USA), 0.5 µL each primers (IDT, Inc.), 0.5 units of Taq polymerase (Promega), 2 µL of total DNA (10 ng/µL). The final volume was adjusted using nuclease free water (Promega, Madison, USA). For the negative control, the DNA template was replaced with deionized water. All the samples were vortexed and the reaction was done in a Veriti 96-Well Thermal Cycler (Veriti® Thermal Cycler, Applied Biosystems, USA). PCR cycling was completed with an initial denaturation at 95°C for 3 min followed by 35 cycles of denaturation at 95°C for 20 s, annealing at 58°C for 20 s and extension at 72°C for 30 s and the final extension was performed at 72°C for 5 min. Amplified PCR target was analyzed in 2% agarose gel in a horizontal electrophoresis chamber (SUB13, Hoefer, Inc., California, USA) with FloroSafe DNA Stain (1st Base Laboratories, Selangor, Malaysia) at a constant voltage of 80 V for 120 min along with 50 bp DNA ladder (Promega, USA). Separated products on agarose-gel were visualized under ultraviolet (UV) transilluminator-gel documentation system (Alpha Imager HP; Alpha InfoTech Corp., San Leandro, CA, USA).
Specificity and Sensitivity Tests
To check the feline specificity of the primer pairs, cross-amplification was performed against DNA templates of 17 non-target species (beef, water buffalo, chicken, goat, sheep, domestic duck, pigeon, turkey, carp, cuttle, tilapia, shrimp, onion, tomato, wheat, rat, and pork). However, the sensitivity of the target DNA was in mixed meats (10, 5, 0.2, and 0.01%) and (10, 5, 1, 0.1, and 0.01%) in cat meat contaminated meatballs. The limit of detection was 0.01% (w/w) feline DNA under mixed-meats and 0.1% (w/w) feline meat in meatball matrices.
Comparison of Target DNA Stability
To study the comparative stability of the newly designed 69 bp and one of the previously documented 108 bp targets (the shortest of the previously documented targets 672, 331, 274, 180, and 108 bp) PCR was performed with 50 ng feline DNA template after autoclaving feline meats at 133°C for 30, 60, 90, 120, and 150 min under 43.51 psi and microwave cooking at 600 and 700 W for 30 min.
Result and Discussion
Commercial kit (Yeastern Biotech Co. Ltd., Taiwan) provided higher yield (120–692 ng/µL) of DNA from raw and treated meat samples than conventional liquid–liquid extraction techniques.[Citation22] Commercial DNA extraction kit was also safe for handling to extract DNA with minimal damage.[Citation22] The concentration and purity of the extracted DNA samples were determined based on spectrophotometric measurements of absorbance at 260 and 280 nm. A260/280 value of all extracted DNA was 1.7 to 2.0 which reflected good purity.[Citation23] Approximately, 1 g admixtures and meatballs were used to extract DNA using the CTAB method. The yield of DNA from treated admixed samples was 550–700 ng/µL, whereas from beef and chicken meatballs was 170–373 and 190–347 ng/µL, respectively. These results were consistent with an earlier report.[Citation19] The lower yield of DNA from meatballs might be due to the presence of plant spices, salts, and oil ingredients which were used to formulate beef and chicken meatballs (). Heat treatment also increased the number of cells in per unit weight of tissue through dehydration.[Citation22]
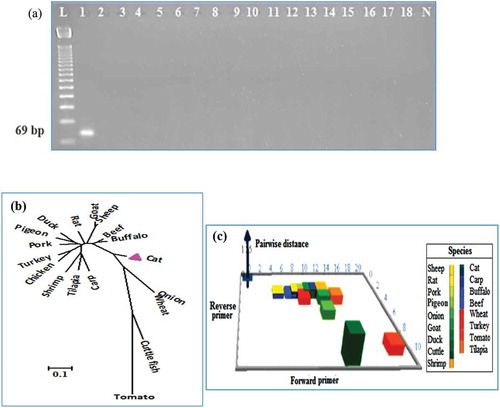
A 69-bp fragment was successfully amplified from 1140-bp cytb gene of feline mitochondria (Accession number: AB194812.1, location 15–83). Such product was never obtained from other 17 species of land, aquatic, and plant origins used in this study. The cytb and cob gene sequences of each species were retrieved from NCBI data base (cat: AB194812.1, beef: EU807948.1, chicken: EU839454.1, domestic duck: HQ122601.1, sheep: EU365990.1, water buffalo: D32193, pig: GU135837.1, turkey: HQ122602.1, goat: EU130780.1, rat: HM222710.1, pigeon: KC811464.1, carp: AB158807.1, cuttle: AB240155.1, tilapia: AF015020, shrimp: EU069446.1, tomato: XM004251454.1, onion: GU253304.1, wheat: X02352.1). We found an optimum primer concentration of 20 picomoles and preheating temperature of 95°C for 3 min, denaturation at 95°C for 20 s in 35 cycles, annealing at 58°C for 20 s, and 30 s of extension at 72°C and 5 min of final extension at 72°C were found suitable for the amplification of desired products. No cross species positive PCR amplification was found in repeated PCR run along with 17 non-target species. A higher annealing temperature increases primer specificity and reduces non-specific PCR amplification.[Citation2,Citation24] The optimized annealing temperature of 58°C was favorable for the detection of 69 bp feline targets in all experimental analysis.
Species-specific PCR assays,[Citation19,Citation25] are widely used to detect a minute level of DNA in raw, processed, and commercial food products because of its rapidity, simplicity, sensibility, and specificity.[Citation26] Multiplex PCR,[Citation27,Citation28] is a most recent development in the field of PCR technology. It is theoretically interesting and can distinguish many species in a single assay platform but the optimization of PCR reactions in a single tube is really an outstanding and challenging job.[Citation1] In contrast, species-specific PCR is simple, precise, and easy to handle without any expensive equipment and chemicals.[Citation29] Mitochondrial (mt-cytb) gene of F. catus was retrieved from NCBI genome sequence database and was used to develop a pair of primers using Primer3Plus software targeting a 69-bp conserved site of the said species. BLAST analysis tool in the NCBI database was used to check the theoretical feline specificity and dissimilarity with other tested and non-tested species (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple alignment analysis using the ClustalW (http://www.genome.jp/tools/clustalw/), showed 100% similarity only with the cytb gene sequence of F. catus and 3–19 nucleotide mismatching with other 17 tested species (). The maximum composite likelihood method,[Citation30] was used to analyze the phylogenetic tree () and pairwise distance () in the primer binding regions of the tested species.[Citation31] The highest pairwise distance (1.29) was observed between cat and cuttle fish and the lowest (0.21) was found with beef and buffalo.[Citation30] Similar results were found when a 3D plot was created among the other species ().[Citation32] All the obtained data represented highly discriminating properties of the designed F. catus specific primers from the other non-target animal, fish and plant species (). It has been reported that a single mismatch in the primer binding region might cause amplification failure in PCR reaction.[Citation2,Citation24] In this assay, the maximum and the minimum mismatches in the primer binding region were (>19 nt) and (>3 nt), respectively. pointed that cross-reaction probability was unlikely in an actual PCR run. The PCR experiment amplified 69-bp target product only from F. catus DNA template (), confirming the in silico or theoretical data analysis. Previously, cat meat was detected by PCR assays targeting mitochondrial whole genome (672 bp);[Citation14] (672 bp);[Citation13] 274 bp (ND4);[Citation15] 108 bp (12S rRNA);[Citation17] 180 bp (cytb);[Citation18] and 331 bp (cytb).[Citation16] However, it is presumed that longer amplicons are thermodynamically less stable than the shorter ones under environmental decomposition or degradation or by food processing treatment.[Citation31,Citation33,Citation34] Since the target of the currently available assays are longer (672–108 bp), there is a strong probability of target fragmentation under thermal and other cooking or processing treatments.[Citation2,Citation33,Citation34] The documented assays also need to be adapted and validated for the analysis of process foods which contain a broad spectrum of matrices that might inhibit a PCR amplification. For the first time, we developed and optimized here a very short length amplicon (69 bp) PCR assay targeting mt-cytb gene for F. catus and demonstrated its stability under various food processing treatments for the analysis of processed foods for feline adulteration detection.
FIGURE 1 Cross-amplification of feline-specific target (69 bp; Lane 1) against 17 different species (Lanes 2–18). A: In A, Lane L: 50 bp ladder, 1: cat, 2: beef, 3: buffalo, 4: chicken, 5: goat, 6: sheep, 7: pigeon, 8: pork, 9: duck, 10: rat, 11: turkey, 12: carp, 13: cuttle, 14: shrimp, 15: tilapia, 16: tomato, 17: onion, 18: wheat, and Lane N: negative control. In B, dendogram built by neighbor-joining method against primer binding region of the feline target and similar region of the cytochrome b/cob gene of the other species said in A. In C, 3D plot showing mismatch and pairwise distance of the forward and reverse primers.
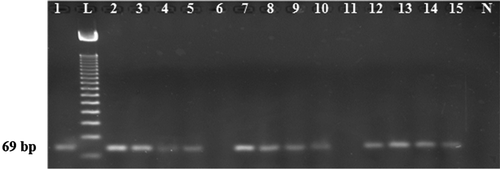
The specificity and sensitivity were checked in ternary mixtures which were composed of beef and wheat flour contaminated with 10, 5, 0.2, and 0.01% (w/w) of cat meat. We demonstrated that F. catus specific PCR assay developed in this study was highly sensitive and specific since it unambiguously identified as low as 0.01% (w/w) feline meat in raw, autoclaved, and boiled cat-beef-wheat flour ternary admixes (). However, the sensitivity of the previously documented assays has not been systematically studied. In addition, Ali et al.,[Citation19] identified up to 0.01% (w/w) canine DNA under mixed matrices (formulation of admixes with target and non-target species). Martin and his co-workers[Citation17] detected 0.1% (w/w) cat meat spiked with binary mixtures (oat:cat) under standard autoclaving condition (133°C for 30 min) with 108 bp target amplicon. However, they did not check its stability under extensive autoclaving (120°C for 2.5 h under 45 psi) and extreme microwaving (700 W for 30 min) which are known to massively degrade target DNA.[Citation31,Citation33] Ilhak and Arslan,[Citation15] identified 0.1% of feline species from beef:goat:lamb ternary mixtures with 274 bp target amplicon. However, they have not tested their assay in any thermal processing condition and commercial meat products. The highest detection limit 0.01% of cat meat, [Citation14] was obtained in a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay that targeted a 672-bp site of mt-cytb gene in ternary mixtures (beef:goat:lamb). Nonetheless, the authors did not state the concentration of template DNA that they used. More importantly, stability of such a long length target (672 bp) under compromised states is seriously questioned.[Citation19] Thus the merit of this newly developed assay over the others is evident since it is well defined, has used the shortest target (69 bp) and achieved high sensitivity (0.01% (w/w) in ternary admixes and 0.1% (w/w) in complex background in meatballs).
FIGURE 2 Feline adulteration detection in feline, bovine, and wheat flour ternary mixtures. Lanes 1, 6, and 11: 100% raw cat meat, beef, and wheat flour, respectively. 10, 5, 0.2, and 0.01% feline meat containing admixtures before any treatments (Lanes 2–5), after extensive autoclaving (120°C for 2.5 h under 45-psi p; Lanes 7–10) and after boiling (Lanes 12–15). Lanes L: ladder and Lane N: negative control.
Three different food processing treatments, namely, boiling, autoclaving, microwave cooking () which may alter the quality of the DNA or lead to the breakdown of the specific target were used to check the effect of different processing conditions on the feline specific DNA target stability.[Citation15,Citation35,Citation36] Microwave cooking was done at three different stages 300, 500, and 700 W for 30 min and boiling was done at 110°C for 90 min. We found here that none of these treatments affected the stability of the PCR amplification of the feline specific target ().These processing treatments were performed by various authors to check the specificity of canine,[Citation19,Citation31] and swine DNA targets.[Citation29] Here we autoclaved meat under three different pasteurization phases (110°C for 2 h at 14.5 psi; 120°C for 50 min at 14.5 psi; 133°C for 20 min at 43.51 psi) according to the European legislation,[Citation37] and obtained PCR products from 10 ng of target DNA (). Even no adverse effects was found when extensive autoclaving (120°C for 2.5 h at 45 psi),[Citation19] were performed (, Lane 4). In addition to these conventional treatments, newly emerging microwave cooking that cooks homogeneously from all directions, did not breakdown the feline DNA target, even under extreme stage of 700 W for 30 min (, Lane 7).
FIGURE 3 Stability of the feline target under autoclaving at 110°C for 2 h (Lane 1); 120°C for 50 min (Lane 2); 133°C for 20 min (Lane 3), and 120°C for 2.5 h (Lane 4); microwave heating at 300 (Lane 5), 500 (Lane 6), and 700 W (Lane 7) for 15–30 min and boiling at 100°C for 90 min (Lane 8). Lane L: ladder and lane N: negative control.
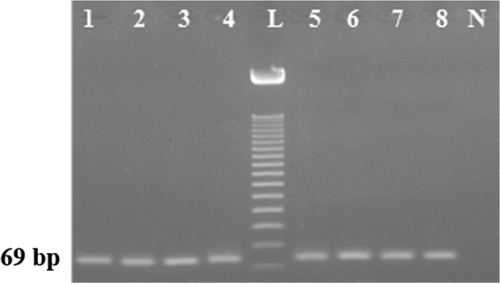
Previously, Abdulmawjood et al.,[Citation14] (672 bp; cytb) and Martin et al.,[Citation17] (108 bp; 12S rRNA) amplified cat meat DNA from raw, boiled, and autoclaved conditions. However, none of them verified their PCR products under extreme autoclaving systems. Here, we studied the target stability under extensive autoclaving and microwaving treatments and found its proven stability under harsh conditions. Our group, Ali et al.[Citation19,Citation34] and Rahman et al.,[Citation31] previously studied the effects of extensive autoclaving treatments on various samples and found short length targets can survive compromised conditions (natural and forceful degradation while cooking). Thus, it was not unpredicted that a 69 bp feline target would be stable under extreme processing treatments.
Minced meats are common ingredients for many commercial food products[Citation37] and meat replacement under mixed and processed conditions has been taken in many places.[Citation8] The “Administration of Czech State Veterinary” discovered horse meat in frozen meatballs marketed as beef and pork meatballs in Sweden,[Citation12] while turkey and chicken was found in 100% beef meatballs in Turkey.[Citation11] Finally, we tested the developed assay in commercial meatballs matrices since no report has been published for feline meat detection in commercial products. To fill up this research gap, different amount (10, 1, 5, 0.1, and 0.01%) of cat meat was spiked in deboned beef and chicken following Rahman et al.[Citation31] To simulate the normal cooking and extensive autoclaving, as prepared meatballs were boiled at 100°C for 90 min and autoclaved at 120°C for 2.5 h under 45 psi, respectively. In ; Lanes (1–4, 6–9, and 11–14) and ; Lanes (7–10 and 12–15) clearly showed that PCR products were obtained from cat meat adulterated beef () and chicken meatballs (). Feline PCR product was amplified from all contaminated meatballs and thus the tested limit of detection (LOD) of the assay was determined to be 0.1% (w/w) feline meats. Previously, Ali et al.[Citation19] and Rahman et al.[Citation31] detected 0.1% (w/w) and 0.2% (w/w) canine DNA in frankfurters and meatball. More importantly, fraudulent replacement of lower valued ingredients with higher valued ingredients is a global issue,[Citation38] therefore, we tested three “halal logo” labeling commercial beef (A, B, C) and chicken (A´, B´, C´) meatballs were collected from different selling spots in Malaysia. Thus, we found a 69 bp feline specific PCR assay was successfully amplified from all cat meat adulterated meatballs but no PCR product was obtained from all commercial meatballs in (Lanes 1–6) and .
TABLE 4 Screening of commercial beef and chicken meatballs
FIGURE 4 Analysis of A: beef and B: chicken meatballs for feline adulteration. In A, Lanes 1–5: raw; 6–10: boiled; and 11–15: autoclaved beef meatballs spiked with 10, 1, 5, 0.1, and 0.01% feline meats, respectively. In B, Lanes 1–3: commercial beef and 4–6: commercial chicken meatballs. Lanes 7–11: boiled and 12–16: autoclaved chicken meatballs spiked with 10, 1, 5, 0.1, and 0.01% feline meats, respectively. Lane L: Ladder and N: negative control.
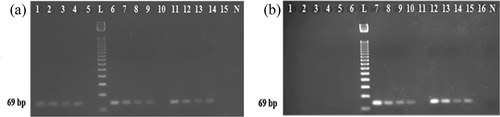
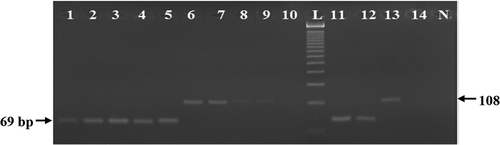
We believe the superiority of a food forensic assay largely depends on its stability, sensitivity, robustness, and precision under various food processing conditions.[Citation13] It has been assumed for long time that short length PCR targets better performs over the longer ones.[Citation19,Citation31,Citation33] However, little scientific efforts have been made to validate or prove this hypothesis. Consequently, we subjected here two different PCR targets (69 bp which was developed in this study and 108 bp that was shortest in length among the published reports) under harsh autoclaving (133°C for 30, 60, 90, 120, and 150 min under 43.51 psi) and microwaving treatments (600 and 700 W for 30 min). We found here that 69 bp target of feline cytb site was stable and, hence were amplified under all treatment conditions (). But 108 bp targets of 12 s rRNA failed to withstand 150 min of autoclaving at 133°C and 30 min of microwaving condition at 700 W (; Lanes 10 and 14). These clearly revealed that the assay we developed here was more stable and robust than those of the previously reported assays.
FIGURE 5 Comparative stability analysis of a shorter (69 bp; developed in this study) (Lanes 1–5 and 11–12) and longer (108 bp; previously documented; Lanes 6–10 and 13– 14 feline DNA targets after autoclaving (lanes 1–10) at 133°C for 30 (Lanes 1 and 6), 60 (Lanes 2 and 7), 90 (Lanes 3 and 8), 120 (Lanes 4 and 9), and 150 (Lanes 5 and 10) min under 43.51 psi and microwave cooking (Lanes 11–14) at 600 W (Lanes 11 and 13) and 700 W (Lanes 12 and 14) for 30 min. Please note that while the 69 bp was amplified under all treatments (Lanes 1–5 and 11–12), the 108 bp target was not detected after 150 min of autoclaving (Lane 10) and microwaving at 700 W for 30 min (Lane 14).
Conclusion
The shortest-amplicon-length (69 bp) PCR assay targeting mitochondrial cytochrome b gene for feline species detection was successfully developed. This shortest-amplicon-length PCR target was more stable over the previously documented PCR biomarkers for feline species detection under autoclaving and microwaving treatments, suggesting its application in food forensic or any archaeological studies. It was tested and optimized under pure, admixed, and commercial food matrices for the sensitive and reliable detection of feline species in raw, admixed, and commercial beef and chicken meatballs. This approach was first for feline material detection in processed foods. The assay amplified the desired target from all matrices without any adverse effects. Specificity was tested by cross-challenging the primers against 17 different species and no cross-species were detected, reflecting the robust and stringent specificity of the primers for feline species. The target was stable under extensive boiling, autoclaving, and microwaving treatments which may cause degradation and breakdown of longer targets. LOD of the assay was 0.01% (w/w) feline meat in admixed meats and 0.1% (w/w) in commercial beef and chicken meatballs. We believed the assay is suitable to be used by regulatory bodies for the routine assessments of feline species in food forensics or archaeological investigations.
Funding
Md. Al Amin is a recipient of Graduate Research Assistantship (GRA) from the University of Malaya, Kuala Lumpur, Malaysia. Md. Al Amin was paid by “Akaun Pengrusan Combicat no. 31377” and “Akaun Penyelidikan Flagship-RU002-2014” to Sharifah Bee Abd Hamid and consumables were paid by GC00114SBS to Md. Eaqub Ali.
Additional information
Funding
References
- Ali, M.E.; Razzak, M.A.; Hamid, S.B.A. Multiplex PCR in Species Authentication: Probability and Prospects—A Review. Food Analytical Methods 2014. DOI:10.1007/s12161-014-9844-4
- Ali, M.E.; Hashim, U.; Mustafa, S.; Che Man, Y.B.; Dhahi, T.S.; Kashif, M.; Uddin, M.K.; Abd Hamid, S.B. Analysis of Pork Adulteration in Commercial Meatballs Targeting Porcine-Specific Mitochondrial Cytochrome b Gene by TaqMan Probe Real-Time Polymerase Chain Reaction. Meat Science 2012a, 91, 454–459.
- Premanandh, J. Horse Meat Scandal–A Wake-Up Call for Regulatory Authorities. Food Control 2013, 34, 568–569.
- Anitei, S. The Origin of SARS Epidemics Found in Civet Cat Meat Consumption in Southern China. Softpedia 2006. http://news.softpedia.com/news/The-Origin-of-SARS-Epidemics-Foundin-Civet-Cat-MeatConsumption-in-Southern-China-40897.shtml (accessed Feb 15, 2015).
- Podberscek, A.L. Good to Pet and Eat: The Keeping and Consuming of Dogs and Cats in South Korea. Journal of Social Issues 2009, 65, 615–632.
- Phillips, T. Chinese Police Find Slaughterhouse Selling Cat Meat. The Telegraph 2013. http://www.telegraph.co.uk/news/worldnews/asia/china/10417032/Chinese-police-findslaughterhouse-selling-cat-meat.html (accessed Feb 15, 2015).
- Chatterji, A. Dog or Cat Meat Suspected in Mystery Indian “Lamb” Curry. International Business Times 2013. http://www.ibtimes.co.uk/dogcat-meat-horse-india-lamb-curry-451343.
- Ali, M.E.; Hashim, U.; Mustafa, S.; Che Man, Y.B. Swine-Specific PCR-RFLP Assay Targeting Mitochondrial Cytochrome b Gene for Semi Quantitative Detection of Pork in Commercial Meat Products. Food Analytical Methods 2012b, 5, 613–623.
- Whitney, T.; Smith, S. Substituting Red Berry Juniper for Oat Hay in Lamb Feedlot Diets: Carcass Characteristics, Adipose Tissue Fatty Acid Composition, and Sensory Panel Traits. Meat Science 2015, 8, 404–407.
- Rohman, A.; Sismindari; Erwanto, Y.; Che Man, Y.B. Analysis of Pork Adulteration in Beef Meatball Using Fourier Transform Infrared (FTIR) Spectroscopy. Meat Science 2011, 88, 91–95.
- Ulca, P.; Balta, H.; Cagin, I.; Senyuva, H.Z. Meat Species Identification and Halal Authentication Using PCR Analysis of Raw and Cooked Traditional Turkish Foods. Meat Science 2013, 94, 280–284.
- Pollak, S. Horsemeat Scandal Spreads to Ikea Swedish Meatballs. Time World 2013. http://world.time.com/2013/02/26/horse meat-scandal-spreads-to-ikea-swedish-meatballs (accessed Feb 15, 2015).
- Abdel-Rahman, S.M.; El-Saadani, M.A.; Ashry, K.M.; Haggag, A.S. Detection of Adulteration and Identification of Cat’s, Dog’s, Donkey’s, and Horse’s Meat Using Species-Specific PCR and PCR-RFLP Techniques. Australian Journal of Basic and Applied Sciences 2009, 3, 1716–1719.
- Abdulmawjood, A.; Schönenbrücher, H.; Bulte, M. Development of a Polymerase Chain Reaction System for the Detection of Dog and Cat Meat in Meat Mixtures and Animal Feed. Journal of Food Science 2003, 68, 1757–1761.
- Ilhak, O.I.; Arslan, A. Identification of Meat Species by Polymerase Chain Reaction (PCR) Techniques. Turkish Journal of Veterinary & Animal Sciences 2007, 31, 159–163.
- Irine, I.; Nuraini, H.; Sumantri, C. Species Authentication of Dog, Cat, and Tiger Using Cytochrome β Gene. Media Peternakan 2013, 36, 171–178.
- Martin, I.; Garcia, T.; Fajardo, V.; Rojas, M.; Hernandez, P.E.; Gonzalez, I.; Martin, R. Technical Note: Detection of Cat, Dog, and Rat Or Mouse Tissues in Food and Animal Feed Using Species-Specific Polymerase Chain Reaction. Journal of Animal Science 2007, 85, 2734–2739.
- Tobe, S.S.; Linacre, A.M. A Multiplex Assay to Identify 18 European Mammal Species from Mixtures Using the Mitochondrial Cytochrome b Gene. Electrophoresis 2008, 29, 340–347.
- Ali, M.E.; Rahman, M.M.; Hamid, S.B.A.; Mustafa, S.; Bhassu, S.; Hasim, U. Canine-Specific PCR Assay Targeting Cytochrome b Gene for the Detection of Dog Meat Adulteration in Commercial Frankfurters. Food Analytical Method 2013. DOI:10.1007/s12161-013-9672-y
- Commission, E. Council Regulation (EC) No 178/2002 of 28 January 2002 Laying Down the General Principles and Requirements of Food Law, Establishing the European Food Safety Authority and Laying Down Procedures in Matters of Food Safety. Off J Euro Comm 2002, L31, 24.
- Ma, X.; Duan, J.; Zhu, D.; Dong, T.; Tsim, K. Species Identification of Radix Astragali (Huangqi) by DNA Sequence of Its 5S-rRNA Spacer Domain. Photochemistry 2000, 54, 363–368.
- Karabasanavar, N.S.; Singh, S.P.; Umapathi, V.; Kumar, D.; Patil, G.; Shebannavar, S.N. A Highly Specific PCR Assay for Identification of Raw and Heat Treated Mutton (Ovis Aries). Small Ruminant Research 2011b, 100, 153–158.
- Adams, D.S. Lab Math: A Handbook of Measurements, Calculations, and Other Quantitative Skills for Use at the Bench, 2nd Ed; Cold Spring Harbor Laboratory Press: New York, NY, 2013; 323.
- Wu, J.H.; Hong, P.Y.; Liu, W.T. Quantitative Effects of Position and Type of Single Mismatch on Single Base Primer Extension. Journal of Microbiological Methods 2009, 77, 267–275.
- Mane, B.G.; Mendiratta, S.K.; Tiwari, A.K. Beef Specific Polymerase Chain Reaction Assay for Authentication of Meat and Meat Products. Food Control 2012, 28, 246–249.
- Mafra, I.; Ferreira, I.M.P.L.V.O.; Oliveira, M.B.P.P. Food Authentication by PCR-Based Methods. European Food Research Technology 2007, 227, 649–665.
- Dalmasso, A.; Fontanella, E.; Piatti, P.; Civera, T.; Rosati, S.; Bottero, M.T. A Multiplex PCR Assay for the Identification of Animal Species in Feedstuffs. Molecular Cell Probes 2004, 18, 81–87.
- Koppel, R.; Ruf, J.; Rentsch, J. Multiplex Real-Time PCR for the Detection and Quantification of DNA from Beef, Pork, Horse, and Sheep. European Food Research Technology 2011, 232, 151–155.
- Karabasanavar, N.S.; Singh, S.; Kumar, D.; Shebannavar, S.N. Detection of Pork Adulteration by Highly-Specific PCR Assay of Mitochondrial D-Loop. Food Chemistry 2014, 145, 530–534.
- Tamura, K.; Nei, M.; Kumar, S. Prospects for Inferring Very Large Phylogenies by Using the Neighbor-Joining Method. Proceedings of the National Academy of Sciences of the United States of America 2004, 101, 11030–11035.
- Rahman, M.M.; Ali, M.E.; Hamid, S.B.A.; Mustafa, S.; Hashim, U.; Hanapi, U.K. Polymerase Chain Reaction Assay Targeting Cytochrome b Gene for the Detection of Dog Meat Adulteration in Meatball Formulation. Meat Science 2014, 97, 404–409.
- Saitou, N.; Ne, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Molecular Biology and Evolution 1987, 4, 406–425.
- Rojas, M.; Gonzalez, I.; Pavon, M.A.; Pegels, N.; Lago, A.; Hernandez, P.E.; Garcia, T.; Martin, R. Novel TaqMan Real-Time Polymerase Chain Reaction Assay for Verifying the Authenticity of Meat and Commercial Meat Products from Game Birds. Food Additives and Contaminants 2010, 27, 749–763.
- Ali, M.E.; Hashim, U.; Dhahi, T.S.; Mustafa, S.; Man, Y.B.C.; Latif, M.A. Analysis of Pork Adulteration in Commercial Burgers Targeting Porcine-Specific Mitochondrial Cytochrome B Gene by TaqMan Probe Real-Time Polymerase Chain Reaction. Food Analytical Methods 2011, 5, 784–794.
- Arslan, A.; Ilhak, O.I.; Calicioglu, M. Effect of Method of Cooking on Identification of Heat Processed Beef Using Polymerase Chain Reaction (PCR) Technique. Meat Science 2006, 72, 326–330.
- Haunshi, S.; Basumatary, R.; Girish, P.; Doley, S.; Bardoloi, R.; Kumar, A. Identification of Chicken, Duck, Pigeon, and Pig Meat by Species-Specific Markers of Mitochondrial Origin. Meat Science 2009, 83, 454–459.
- Tanabe, S.; Hase, M.; Yano, T.; Sato, M.; Fujimura, T.; Akiyama, H. A Real-Time Quantitative PCR Detection Method for Pork, Chicken, Beef, Mutton, and Horse Flesh in Foods. Bioscience Biotechnology Biochemistry 2007, 71, 3131–3135.
- Nurrulhidayah, A.F.; Che Man, Y.B.; Amin, I.; Arieff, S.R.; Farawahidah, M.Y.; Shuhaimi, M.; Khatib, A. FTIR-ATR Spectroscopy Based Metabolite Fingerprinting As a Direct Determination of Butter Adulterated with Lard. International Journal of Food Properties 2015, 18, 372–379.
