Abstract
Pectinase is widely used in the fruit juice industry with various aspects, especially to reduce blur in fruit juice. To produce pectinase, Bacillus subtilis were isolated from Kilis soil contaminated by agricultural crop wastes. The extracellular pectinase was purified by using three different technics (cell-free supernatant, ethanol, and ammonium sulphate precipitation) and enzyme activity was measured with spectrophotometric analysis. The maximum specific activity was observed in enzyme preparate purified by ethanol precipitation, 217.44 U/mg. The degradation products of reaction–glucose, saccharose, and fructose–were identified by Fourier transform infrared spectroscopy and thin layer chromatography analysis. Two protein bands belong to pectinase of B. subtilis were determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Molecular weights of bands were approximately calculated as 60 and 64 kDa.
INTRODUCTION
Pectin, which is a structural polysaccharide and cementing material between adjacent cells for the firmness and structure of the plant cellulose network, presents in its highest concentration in the middle lamella and cell wall of higher plants.[Citation1–Citation6] It is also a polymeric material containing carbohydrate groups that are esterified with methanol.[Citation3–Citation5]
Pectinases (pectinolytic enzymes) consist of a mixture of complex enzymes that catalyze specifically the hydrolysis of pectin-including substrates.[Citation6–Citation8] These enzymes divide into three main classes that catalyze depolymerization, demethylation, and de-esterification reactions.[Citation9,Citation10] Polygalacturonase (EC 3.2.1) that catalyzes the breakdown of α 1-4 glycosidic bonds between two galacturonic acid residues in the smooth region of the pectin molecule is a depolymerizing enzyme.[Citation10,Citation11] Pectin lyase (EC 4.2.2) and pectin esterase (EC 3.1.1) that act as the elimination reaction between two methylated residues and the splitting of methoxyl groups releasing methanol, are demethylating and de-esterifying enzymes, respectively.[Citation10,Citation11]
Pectinases constitute approximately 10% of the total enzyme production in the world market and 25% of global sale in the food industry.[Citation2–Citation4,Citation12] They have been widely used in the fruit juice and textile industry, the wine production industry, the vegetable waste treatment, the pulp and papermaking processes, the degumming of plant fibers, the oil extraction processes, and for the chocolate, tea, and coffee fermentations.[Citation2,Citation5–Citation7,Citation13]
In the fruit juice industry, they are used for clarification of fruit juice by reduction in viscosity, increasing of juice yield by enzymatic liquefaction, and maceration of pulps, enhancing pigmentation by extracting more anthocyanin.[Citation7,Citation9,Citation14] Pectinases are synthesized by different sources, such as bacteria, fungi, yeasts, insects, nematodes, plants, and protozoan.[Citation1,Citation4,Citation6,Citation12] In recent years, microbial pectinases have been widely studied due to the requirement of highly productive strains and the cost-effective production of enzymes. In the present study, the pectinase was purified from a newly isolated Bacillus subtilis strain and this enzyme was characterized.
MATERIAL AND METHODS
Isolation of Microorganisms
Bacterial isolates were obtained from a soil sample contaminated by agricultural crop wastes, Kilis. One gram of the soil sample was homogenized in 9 mL of physiological saline. Then, 100 µL aliquot’s from this homogenization was spread onto pectinase screening plates including (g/L) ammonium sulfate 2, yeast extract 1, Na2HPO4 6, KH2PO4 3, pectin 5, agar 20 (pH 7.0),[Citation12] and plates were incubated at 37°C for 24–48 h.
Screening of Pectinase Producing Bacteria by Plate Assay
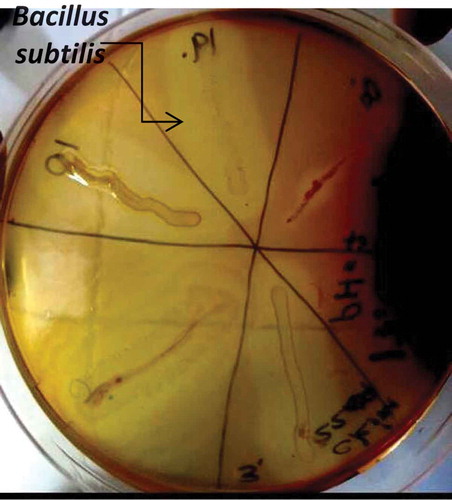
After colonization, the plates were flooded with iodine-potassium iodide solution (1 g iodine, 5 g KI for 330 mL distilled water) and incubated at 37°C for 15 min.[Citation1,Citation10] Following incubation, the plates were investigated for the determination of pectin hydrolysis zones. The isolates observed a clearance zone around their colony that was selected and maintained at 4°C for enzyme purification assays.
Pectinase Fermentation Medium
The strain was growth for pectinase production in liquid fermentation medium containing (g/L) KH2PO4 4, Na2HPO4 0,2, FeSO4.7H20 0,02, CaCl2 0,01 (NH4)2SO4 2, pectin 15 (mg/L) MnSO4.7H20 70, H3BO3 10[Citation15] (pH 7.0), at 37°C for 3 days under agitation (150 rpm).
Partial Purification of Pectinase from Bacillus subtilis
To obtain a crude enzyme, the fermentation medium was centrifuged at 7454 × g for 30 min. The cell-free supernatant was evaluated as enzyme preparate for purification steps which will be described below. A sample from the enzyme constituent was precipitated by adding ethanol 90% of the total volume to supernatant. The mixture was kept at –20°C for 24 h. After precipitation, the suspension was centrifuged at 7454 × g for 30 min to separate into extracellular pectinase. The obtained pellet was dissolved in 0.2 M phosphate buffer, pH 7.0.
The other part of supernatant was saturated by 65% ammonium sulphate with gentle agitation at 4°C for 24 h. After centrifugation, under the same condition, the precipitate was suspended in a phosphate buffer “pH 7.0” and dialyzed at 4°C for 24 h against the same buffer. It was performed enzyme activity and total protein amount assays for each of these crude enzyme samples.
Pectinase Enzyme Activity
A sample of 0.5 mL crude enzyme suspension was incubated with 0.5 mL of 1% pectin substrate (in 0.1 M acetate buffer “pH 6.0”) at 40ºC for 10 min. Then, 1 mL DNS reagent (3,5-dinitrosalicylic 10.6 g, NaOH 19.8 g for 1416 mL distilled water)[Citation16] was added to reaction mixture and this mixture was boiled at 90°C for 5 min. The reaction was finished by adding 1 mL of 1 M Rochelle’s (Seignette) salt. Thereafter, the absorbance of the diluted mixture by adding 2 mL distilled water was spectrophotometrically measured at 595 nm by estimation of reducing sugars.[Citation1,Citation6,Citation12] An isolation and fermentation medium, pectin was used for screening of isolates, for production pectinase, and for enzyme assay.
The possible reaction products by degradation of pectin by a pectinase are soluble reducing sugars. In this analysis, glucose was used as standard reducing sugar. For the calculation of the reducing sugars released by the enzymatic activity, a standard curve was prepared by using standard glucose solution in different concentrations. One unit of pectinase activity was described as 1 µmol glucose that was liberated per min.[Citation1,Citation10,Citation12]
Quantitative Assay of Protein Concentration
The total protein amount in enzyme samples was detected by the method of Lowry, using bovine serum albumin (BSA) as standard.[Citation17]
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) Analysis
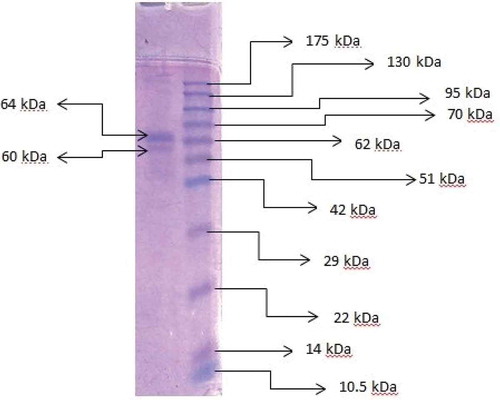
The molecular weight of pectinase that purified from Bacillus subtilis was determined by the Laemmli method using a vertical electrophoresis system including 12% separating and 5% stacking gel.[Citation18] Applied biological material (ABM) opti-protein G252 was used as reference marker protein. Protein bands were examined by staining Coomassie Brilliant Blue R-250.
Fourier Transform Infrared (FTIR) and Thin Layer Chromatography (TLC)
After the hydrolization of pectin by pectinase, the reaction products were identified by FTIR and TLC analysis. First, according to characteristic C-O and C-C stretching vibrations of metabolites, the released sugars were determined by FTIR analysis. FTIR analysis was performed by using Thermo Scientific Nicolet iS10 FTIR Spectrometer. The infrared spectrum was obtained by using attenuated total reflectance (ATR) technic at scan range 4000–650 cm–1. Then the identification of products was continued by TLC analysis. In this analysis, silica gels 60 F254 purchased from Merck were used. The prepared silica gels was activated at 110–120°C for 1 h. Reductive sugars were separated in a solvent system including butanol/acetic acid/water (3:1:1). After separation, the surface of TLC sheets was coated by 25% (v/v) H2SO4 in methanol and was treated at 120°C for 30–45 min.[Citation19] Rf values of the spots belonging to sugars were detected by comparison those of the sugar standards (1%).
RESULTS AND DISCUSSION
Selection of Strain That Showed Pectinolytic Activity in Agar Plate
Pectinolytic activity of bacteria on solid medium including pectin as the only carbon source was examined. As a result of pectin depolymerization, test bacteria that showed clear zone around colonies were evaluated as pectinase producer strains. Accordingly, four bacterial strains were isolated from agar plates (). The strain that demonstrated the largest hydrolysis zone around its colony was revealed to have high pectinase activity, compared to the other strains. The extracellular pectinase purification studies were continued with this strain. This strain was defined as Bacillus subtilis using VITEK bacterial identification system.
Pectinase Activity
The pectinase activity of Bacillus subtilis was performed in extracellular enzyme suspensions from obtained by three different purification steps. Enzyme unit, total protein, and specific activity were given for all crude enzyme preparates in . Based on the incubation period at 37°C for 3 days, the specific pectinase activity was calculated as 62.18 U/mg for supernatant, 217.44 U/mg for ethanol precipitation, and 41.69 U/mg for ammonium sulphate precipitation. The maximum yield of pectinase was acquired by ethanol precipitation. The maximum total activity was calculated as 95.15 U/mL/min for supernatant.
TABLE 1 Enzyme unit, total protein, and specific activity for all enzyme preparates
As indicated in previous studies, the production of extracellular pectinase from Bacillus strains was investigated.[Citation1,Citation8,Citation20,Citation21] The total activity of pectinolytic enzyme belonging to Bacillus sp. RCPTM1 isolated from carrot waste was reported as 49.58 U/mL.[Citation22] In another study, pectinolytic activity for Bacillus polymyxa NCIM 2534 strain was found as 67.33 U/mL.[Citation23] The total activity of pectinase produced by Bacillus firmus within a fermentation time of 18 h was recorded as 56 U/mL.[Citation24] In another study, the specific and total activity of pectinase enzyme purified from Bacillus sp. SC-H strain that showed that pectinase activity at low temperatures were reported as 0.18 U/mL and 7.04 U/mg at 30°C, respectively.[Citation25] Similar results for Bacillus circulans were stated by Raju and Divakar.[Citation4] In their studies, the maximum pectinase production was obtained with submerged fermentation media including galactose (256 U/mL). The cell-free supernatant of Bacillus pumilus strain EK-17 for pectinolytic activities was used and the maximum specific activity was detected as 41 IU/g in the presence of pectin at pH 8.0.[Citation26] The maximum total activity of pectinase from Bacillus cereus grown in medium including pectin as sole carbon source at 37°C was notified as 44 U/mL.[Citation27] The maximum specific activity of pectinase purificated from B. subtilis by centrifuged and ion exchange chromatography was declared as 3.1 and 4.5 U/mg, respectively.[Citation28] Compared to the literature, it is clear that extracellular pectinase obtained from B. subtilis demonstrates high activity and has the potential to be used as an agent for clarification of fruit juice after careful examinations.
Characterization of Pectinase by SDS-PAGE Analysis
For SDS-PAGE, the enzyme preparate purified by ethanol precipitation with the maximum specific activity, was used. According to the SDS-PAGE analysis, two protein bands located at approximately 60 and 64 kDa for extracellular pectinase were shown (). Molecular weights of the partial purified pectinase from various bacteria species were determined as follows: 37 kDa for Paenibacillus xylanolyticus,[Citation29] 89 kDa for Bacillus cereus NRC20,[Citation30] 31 kDa for Streptomyces sp. GHBA10,[Citation31] 37 kDa for Bacillus sp. MFW7,[Citation12] 106 kDa Bacillus sp. DT7,[Citation8] and 66 kDa for Bacillus sp. MBRL576.[Citation32]
Identification of Reducing Sugars by FTIR and TLC
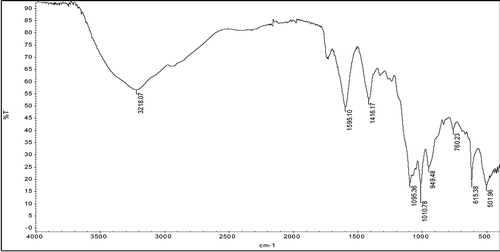
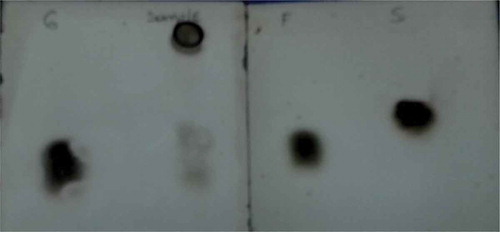
For identification of reducing sugars as degradation products, FTIR and TLC analysis were performed. The most intense peaks belonging to standard sugars were characterized to the C-O and C-C stretching vibrations in the FTIR spectrum.[Citation33] These specific peaks for glucose, fructose, and saccharose appeared at 1033, 1063, and 995 cm–1, respectively. The results of this study were taken as a reference for our FTIR analysis. Accordingly, in the FTIR analysis of reaction mixture, characteristic C-O and C-C stretching vibrations for glucose (at 1010.78 cm–1), fructose (at 1095.36 cm–1), and sucrose (at 949.48 cm–1) were identified (). The wave numbers of the characteristic peaks in FTIR spectrum were seen to shift to higher/lower wavelengths. These shifts in peaks can be clarified with the presence of some impurities in sample. It was detected that glucose, fructose, and saccharose standards migrated with Rf values 0.29, 0.27, and 0.20 on TLC plates. In comparison with the spots of standards, only one spot, at Rf value 0.30, belonging to glucose was observed (). The lack of spots relating to the other sugars can be explained because that glucose concentration was higher than fructose and saccharose in the reaction mixture. In FTIR and TLC analysis, the observation of reducing sugars indicates the degradation of pectin polymer by pectinase during fermentation.
CONCLUSION
In the present study, the extracellular pectinase was produced by using Bacillus subtilis isolated from agricultural crop wastes. The maximum specific activity of this enzyme partially purificated by three different steps, was obtained by ethanol precipitation (217.44 U/mg). During fermentation including pectin as a substrate, the results of FTIR and TLC showed that the removal of pectin will reduce sugars as glucose, fructose, and saccharose. Therefore, these data indicate that it is a better source for various industrial areas, such as textile, food, plant tissue maceration, and fruit juice wastewater treatment.
ACKNOWLEDGMENTS
This study was supported by Kilis 7 Aralık University Research Fund.
REFERENCES
- Karthik, J.L.; Kumar, G.; Rao, K.V.B. Screening of il. Asian Journal of BiochPectinase Producing Microorganisms from Agricultural Waste Dump Soemical and Pharmaceutical Research 2011, 2, 329–337.
- Naderi, S.; Naghavi, N.S.; Shahanipoor, K. Pectinolytic Activity of Aspergillus Niger on Pectic Agricultural Substrates. The 1st International and the 4th National Congress on Recycling of Organic Waste in Agriculture, Isfahan, Iran, Apr 26–27, 2012; pp. 1–6.
- Pedrolli, D.B.; Monteiro, A.C.; Gomes, E.; Carmona, E.C. Pectin and Pectinases: Production, Characterization, and Industrial Application of Microbial Pectinolytic Enzymes. The Open Biotechnology Journal 2009, 3, 9–18.
- Raju, E.V.N.; Divakar, G. Screening and Isolation of Pectinase Producing Bacteria from Various Regions in Bangalore. International Journal of Research in Pharmaceutical and Biomedical Sciences 2013, 4, 151–154.
- Sittidilokratna, C.; Suthirawut, S.; Chitradon, L.; Punsuvon, V.; Vaithanomsat, P.; Siriacha, P. Screening of Pectinase Producing Bacteria and Their Efficiency in Biopulping of Paper Mulberry Bark. Science Asia 2007, 33, 131–135.
- Torimiro, N.; Okonji, R.E. A Comparative Study of Pectinolytic Enzyme Production by Bacillus Species. African Journal of Biotechnology 2013, 12, 6498–6503.
- Geetha, M.; Saranraj, P.; Mahalakshmi, S.; Reetha, D. Screening of Pectinase Producing Bacteria and Fungi for Its Pectinolytic Activity Using Fruit Wastes. International Journal of Biochemistry and Biotechnology Science 2012, 1, 30–42.
- Kashyap, D.R.; Chandra, S.; Kaul, A.; Tewari, R. Production, Purification, and Characterization of Pectinase from a Bacillus sp. DT7. World Journal of Microbiology and Biotechnology 2000, 16, 277–282.
- Bayoumi, R.A.; Yassin, H.M.; Swelim, M.A.; Abdel-All, E.Z. Production of Bacterial Pectinase(S) from Agro-Industrial Wastes Under Solid State Fermentation Conditions. Journal of Applied Sciences Research 2008, 4, 1708–1721.
- Soares, M.M.C.N.; Da Silva, R.; Gomes, E. Screening of Bacterial Strains for Pectinolytic Activity: Characterization of the Polygalacturonase Produced by Bacillus Sp. Revista de Microbiologia 1999, 30, 299–303.
- Prathyusha, K.; Suneetha, V. Bacterial Pectinases and Their Potent Biotechnological Application in Fruit Processing/Juice Production Industry: A Review. Journal of Phytology 2011, 3, 16–19.
- Mukesh Kumar, D.J.; Saranya, G.M.; Suresh, K.; Andal Priyadharshini, D.; Rajakumar, R.; Kalaichelvan, P.T. Production and Optimization of Pectinase from Bacillus sp. MFW7 Using Cassava Waste. Asian Journal of Plant Science and Research 2012, 2, 369–375.
- Zohdi, N.K.; Amid, M. Optimization of Extraction of Novel Pectinase Enzyme Discovered in Red Pitaya (Hylocereus Polyrhizus) Peel. Molecules 2013, 18, 14366–14380.
- Chauhan, S.K.; Tyagi, S.M.; Singh, D. Pectinolytic Liquefaction of Apricot, Plum, and Mango Pulps for Juice Extraction. International Journal of Food Properties 2007, 4, 103–109.
- Maldonada, M.C.; Strasser de Saad, A.M.; Callieri, D. Purification and Characterization of Pectinesterase Produced by a Strain of Aspergillus Niger. Current Microbiology 1998, 28, 193–196.
- Adney, B.; Baker, J. Measurement of Cellulase Activities. Laboratory Analytical Procedure (LAP), Technical Report NREL/TP-510-42628 1996; pp. 1–13.
- Lowry, O.H.; Rosgrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. The Journal of Biological Chemistry 1951, 193, 165–275.
- Laemmli, U.K. Cleavage of the Structural Proteins During Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685.
- Nayebyazdi, N.; Tajick Ghanbary, M.A. Pectinase Activity of Some Micromycetes Isolated from Agricultural Soils. Journal of Sciences Islamic Republic of Iran 2012, 23, 305–311.
- Ouattara, H.G.; Koffi, B.L.; Karou, G.T.; Sangaré, A.; Niamke, S.L.; Diopoh, J.K. Implication of Bacillus sp. in the Production of Pectinolytic Enzymes During Cocoa Fermentation. World Journal of Microbiology and Biotechnology 2008, 24, 1753–1760.
- Ahlawat, S.; Mandhan, R.P.; Dhiman, S.S.; Kumar, R.; Sharma, J. Potential Application of Alkaline Pectinase from Bacillus Subtilis SS in Pulp and Paper Industry. Applied Biochemistry and Biotechnology 2008, 149, 287–293.
- Patil, R.C.; Murugkar, T.P.; Shaikh, S.A. Extraction of Pectinase from Pectinolytic Bacteria Isolated from Carrot Waste. International Journal of Pharma and Bio Sciences 2012, 3, 261–266.
- Singh, M.P.N.; Sinha, M.P. Effect of Cellulose on Secretion of Pectolytic and Cellulolytic Enzymes by Blight Pathogens. Asian Journal of Microbiology 2001, 3, 311–314.
- Roosdiana, A.; Prasetyawan, S.; Mahdi, C.; Sutrisno, S. Production and Characterization of Bacillus Firmus Pectinase. The Journal of Pure and Applied Chemistry Research 2013, 2, 35–41.
- Cabeza, M.S.; Baca, F.L.; Puntes, E.M.; Loto, F.; Baigorí, M.D.; Morata, V.I. Selection of Psychrotolerant Microorganisms Producing Cold-Active Pectinases for Biotechnological Processes at Low Temperature. Food Technology and Biotechnology 2011, 49, 187–195.
- Das, B.; Chakraborty, A.; Ghosh, S.; Chakrabarti, K. Studies on the Effect of pH and Carbon Sources on Enzyme Activities of Some Pectinolytic Bacteria Isolated from Jute Retting Water. Turkish Journal of Biology 2011, 35, 671–678.
- Namasivayam, E.; Ravindar, J.D.; Mariappan, K.; Jiji, A.; Kumar, M.; Jayaraj, R.L. Production of Extracellular Pectinase by Bacillus Cereus Isolated from Market Solid Waste. Journal of Bioanalysis and Biomedicine 2011, 3, 70–75.
- Gyan, D.T.; Javed, Z.; Adarsh, K.S. Pectinase Production and Purification from Bacillus Subtilis Isolated from Soil. Advances in Applied Science Research 2014, 5, 103–105.
- Giacobbe, S.; Pepe, O.; Ventorino, V.; Birolo, L.; Vinciguerra, R.; Faraco, V. Identification and Characterization of a Pectinolytic Enzyme from Paenibacillus Xylanolyticus. BioResources 2014, 9, 4873–4887.
- Ashour, S.M.; Kheiralla, Z.M.H.; Eldiwany, A.I.; Maany, D.A. Production, Purification, and Characterization of Polysaccharide Lytic Enzymes of a Marine Isolate, Bacillus Cereus NRC-20 and Their Application in Biofilm Removal. African Journal of Microbiology Research 2014, 8, 2492–2504.
- Arijit, D.; Sourav, B.; Naimisha, R.V.; Rajan, S.S. Improved Production and Purification of Pectinase from Streptomyces sp. GHBA10 Isolated from Valapattanam Mangrove Habitat, Kerala, India. International Research Journal of Biological Sciences 2013, 2, 16–22.
- Bhardwaj, V.; Garg, N. Production, Purification of Pectinase from Bacillus sp. MBRL576 Isolate and Its Application in Extraction of Juice. International Journal of Science and Research 2012, 3, 648–652.
- Chis, A.; Fetea, F.; Matei, H.; Socaciu, C. Evaluation of Hydrolytic Activity of Different Pectinases on Sugar Beet (Beta Vulgaris) Substrate Using FT-MIR Spectroscopy. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 2011, 39, 99–104.