ABSTRACT
56 lactic acid bacteria were isolated during shalgam fermentation and identified as Lactobacillus spp. (51 isolates), Lactococcus spp. (3 isolates), Streptococcus sp. (one isolate), and Leuconostoc sp. (one isolate). 53 of all isolates decarboxylated both arginine and tyrosine, while others decarboxylated one of arginine or tyrosine. None of the isolates could decarboxylate histidine, ornithine, lysine, phenylalanine, or tryptophan. All isolates produced both agmatine (105.8–867.5 mg L–1) and tyramine (24.5–649.7 mg L–1). Although none of the isolates displayed ornithine decarboxylase activity, putrescine was produced (2.1–33.3 mg L–1) by all isolates, except one Lactobacillus strain. Therefore, lactic acid bacteria seem to be responsible mainly for tyramine and agmatine formation during shalgam fermentation, as well as a small amount of putrescine.
Introduction
Shalgam is a traditional Turkish fermented beverage which has a sour taste with a turbid and red colored appearance. It is produced by lactic acid fermentation and the major raw material is black carrot. The other ingredients are turnip, rock salt, sourdough, bulgur flour, and drinkable water.[Citation1,Citation2] “Kanji,” a traditional beverage in India, and “lactofermented carrot juice” are similar products to shalgam.[Citation1,Citation3] Although there is limited data on microbiology of shalgam, it is known that Lactobacillus species are the dominant lactic acid bacteria (LAB) in traditionally fermented shalgam.[Citation4] Recently, Lactobacillus plantarum and Lactobacillus paracasei subsp. paracasei have been determined as the main species during shalgam fermentation.[Citation5]
In metabolisms of LAB amino acids play many significant roles such as intracellular pH control, generation of metabolic energy or redox power, and resistance to stress.[Citation6] Amino acid metabolism of LAB is also important for food safety and quality. For instance, some amino acids in certain foods contribute to formation of characteristic aroma, while some of them are precursors of biogenic amines.[Citation7–Citation12]
Biogenic amines formed by decarboxylation of amino acids may be found in many foods including fermented foods such as cheese, wine, and beer. In other words, biogenic amines can be formed in foods which contain free amino acids and certain microorganisms capable of decarboxylase activity.[Citation13] Although low levels of biogenic amines can be tolerated, intake of these compounds at high levels causes intoxication.[Citation14] The health problems regarding high intake of biogenic amines include headache, respiratory distress, cardiac palpitation, hypertension or hypotension, facial flushing, itching, swelling, diarrhea, vomiting, migraine headache, and several allergy-related disorders, moreover anaphylactic shock syndrome and death.[Citation13–Citation15]
Considering the LAB are potential biogenic amine producer and shalgam is a beverage produced by lactic acid fermentation, biogenic amine formation in shalgam is expected. The prevailing biogenic amine in shalgam has been reported as putrescine by Özdestan and Üren.[Citation16] These researchers also determined the other biogenic amines including tyramine, agmatine, tryptamine, 2-phenylethylamine, spermine, spermidine, and histamine. The aims of this study were to isolate LAB during fermentation of traditionally produced shalgam and to determine their decarboxylase activities by amino acid decarboxylase test and by screening the biogenic amines using high-performance liquid chromatography (HPLC).
Materials and methods
Production of shalgam
The traditional method was used for shalgam production.[Citation17] One hundred grams of black carrot, 50 g bulgur flour, 7.5 g sourdough, 10 g turnip, and 7.5 g salt were used to obtain 500 mL of shalgam juice. Bulgur flour, sourdough, salt (2.5 g) and 110 mL water were mixed, and then the mixture was fermented at 25°C for 3 days. The fermented mixture was extracted by adding appropriate amount of water and stirring for 5–10 min. This procedure was repeated four times. Extract was filtered and transferred into a glass jar. Then, the chopped black carrot, turnip, and the remaining portion of salt were added to extract and the volume was completed to 500 mL with water. This mixture with black carrot and turnip was fermented at 25°C for 5 days. The pH value reduced from 4.0 ± 0.1 to 3.5 ± 0.0, while the titratable acidity increased from 1.1 ± 0.2 to 4.5 ± 0.3 g L–1.
Isolation and identification of LAB
At each day of fermentation one glass jar was opened for sampling and serial dilutions were prepared with sterile physiological saline solution (0.85% NaCl). Then, 0.1 mL of dilutions were spread on De Man Rogosa Sharp (MRS) Agar (Merck, Darmstadt, Germany) and plates were incubated microaerobically at 30°C for 48 h. After incubation white colonies were picked and grown in MRS broth (Merck, Darmstadt, Germany) at 30°C for 48 h. Isolates were maintained in MRS broth with glycerol (15%) at –20°C. Gram and catalase reactions of isolates were determined initially. Then the gram-positive and catalase-negative isolates were identified by the tests of gas production from glucose, arginine hydrolysis, and growth at different temperatures (10, 15, and 45°C) using the identification scheme recommended by Schillinger and Lücke.[Citation18]
Determination of amino acid decarboxylase activity
Modified decarboxylase medium described by Maijala[Citation19] was used for determination of decarboxylase activity of LAB isolates. Five grams tryptone, 8 g meat extract, 4 g yeast extract, 0.5 g tween-80, 0.2 g MgSO4, 0.05 g MnSO4, 0.04 g FeSO4, 0.1 g CaCO3, and 0.06 g bromocresol purple were dissolved in 1 L distilled water and autoclaved (10 min at 121°C). The corresponding amino acids (L-histidine monohydrochloride, L-tyrosine, L-ornithine monohydrochloride, L-arginine monohydrochloride, L-lysine monohydrochloride, L-tryptophan, and L-phenylalanine [Merck, Darmstadt, Germany]) were added individually to medium at a 0.5% final concentration, and purple bromocresol as pH indicator. By this way, an individual MRS broth tube was prepared for each amino acid. The pH was adjusted to 5.3 aseptically.
MRS broth cultures (30°C, 48 h) were inoculated into modified decarboxylase medium without amino acids and tubes were incubated at 30°C for 5 days. A volume of 0.2 mL of each culture was inoculated into 2 mL of the same medium with corresponding amino acids. The modified decarboxylase medium without amino acid was used as control. One milliliter of sterile liquid paraffin was added into the tubes to obtain anaerobic conditions, and then the tubes were incubated at 30°C for 3 days. The conversion of the color to purple in amino acid added tubes while to yellow in control tubes was evaluated as amino acid decarboxylase positive. Thus, any other result was evaluated as amino acid decarboxylase negative.
Determination of biogenic amine production
LAB isolates were propagated in MRS broth. Each isolate was inoculated (0.2 mL) into 15 mL MRS broth contained arginine (0.2%) and tyrosine (0.2%). Following incubation at 30°C for 5 days, cultures were derivatized and biogenic amines were determined quantitatively by HPLC.[Citation19,Citation20]
Biogenic amine analyses by HPLC
Reagents
Agmatine, putrescine, and tyramine standards as hydrochloride salts, dansylchloride, and Na-glutaminate were obtained from Sigma Chemical (St. Louis, Missouri, USA); perchloric acid, sodium carbonate, trichloroacetic acid, acetonitrile, and acetone from Merck (Darmstadt, Germany). Biogenic amine standard solutions were prepared in 0.4 M perchloric acid to a final concentration of 0.2 mg L–1 for each biogenic amine. Dansyl chloride solution was prepared by dissolving 100 mg dansyl chloride in 10 mL acetone. Two grams of sodium carbonate (Na2CO3) was dissolved in 10 mL and 200 mg sodium glutaminate in 4 mL deionized water.
Chromatographic system and conditions
HPLC determinations were performed with Lachrom Elite (Hitachi, Tokyo, Japan) HPLC system equipped with L-2300 column oven (20°C), L-2130 HTA pump, L-2200 autosampler with a 25 µL loop and L-2455 diode array detector. Detection was performed at 254 nm. Kromasil 100 C18 (100 × 4.6 mm, 3.5 µm) chromatographic column was used.
The buffer (pH 8) consisted of 0.1 M tris-(hydroxymethyl)-aminomethan (40%, Sigma), 0.1 M acetic acid (20%, Merck) and water (40%). Solvent A was prepared using 30 mL of buffer, 550 mL acetonitrile (Merck) and 420 mL water. Solvent B was a mixture of 2 mL of buffer, 900 mL acetonitrile, and 100 mL of water. An initial linear gradient elution program was used to separate amines.
Derivatization
A 10 mL portion of LAB culture grown in MRS broth added tyrosine and arginine was mixed with 25 mL of perchloric acid (0.4 M) and shaken vigorously for 1 h. The solution was diluted to 50 mL by adding trichloroacetic acid (5%). From this solution, a 400 µL portion was mixed with 400 µL Na2CO3 and 400 µL dansyl chloride and held in water bath at 40°C for 30 min. Two hundred microliters of Na-glutaminate was added to the mixture, and after vortexing it was held at 40°C for 60 min. Then, 1 mL of acetonitrile was added to the tube. Following centrifugation at 1191 × g for 20 min, the supernatant was removed, filtrated (0.22 µm), and maintained at –20°C.[Citation21] The analytical determinations were done in duplicate.
Results and discussion
Eighty-five colonies were picked from MRS agar plates and 56 of them, which were gram-positive and catalase-negative, were confirmed as LAB (). Of the 56 LAB isolates, 51 were identified as Lactobacillus spp. and three were Lactococcus spp. Two isolates were identified as Streptococcus sp. and Leuconostoc sp. These findings are in accord with the results of previous studies,[Citation4,Citation5,Citation22] those reported that the Lactobacillus species were dominant during shalgam fermentation. Correspondingly, the presence of Lactococcus and Leuconostoc species in shalgam microflora was previously reported.[Citation2,Citation5,Citation22]
Table 1. Amino acid decarboxylase activity test results of lactic acid bacteria isolated from shalgam.
Fifty-three isolates decarboxylated both arginine and tyrosine. Lactobacillus sp. 4C10 and Leuconostoc sp. 4E9 showed only tyrosine decarboxylase activity, while Lactobacillus sp. 2C5 showed only arginine decarboxylase activity (). The LAB isolates in this study did not decarboxylate histidine, ornithine, lysine, phenylalanine or tryptophan in amino acid decarboxylase medium. Although histidine decarboxylase activity has been shown in some Lactobacillus, Oenococcus, Pediococcus, and Tetragenococcus strains,[Citation23–Citation25] histamine production has not been proposed as common in LAB strains. Similar to our results, Bover-Cid et al.[Citation26] and Moreno-Arribas et al.[Citation10] reported that none of LAB isolates they tested produced histamine. Regarding lysine and ornithine decarboxylation, it has been stated that this kind of metabolism is very rare in LAB.[Citation27] Also, phenylalanine and tryptophan decarboxylation activities are known as uncommon in LAB as reported by Bover-Cid et al.,[Citation28] who stated that phenylethylamine and tryptamine formations by Lactobacillus strains from precursor amino acids were not determined except L. curvatus strains.
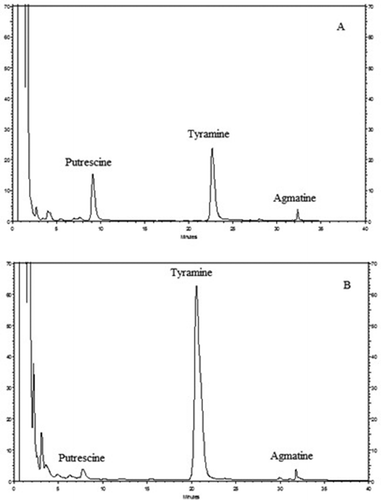
Considering all of the isolates decarboxylated arginine and/or tyrosine; agmatine, putrescine and tyramine were screened quantitatively in amino acid added MRS broth cultures of these isolates. shows HPLC chromatograms of biogenic amine standards and a LAB strain. All of the isolates produced agmatine, tyramine, and putrescine, except one Lactobacillus sp. (0A8) that produced agmatine and tyramine but not putrescine. Agmatine amounts were between 105.8 ± 14.0 and 867.5 ± 21.9 mg L–1, while tyramine amounts were ranged from 24.5 ± 6.9 to 649.7 ± 55.1 mg L–1 (). However, putrescine levels were much lower than both agmatine and tyramine, those were between 2.1 and 33.3 ± 1.7 mg L–1. Although Lactobacillus sp. 2C5 gave negative result in the tyrosine decarboxylase medium, it produced 225.7 ± 76.4 mg L–1 tyramine in tyrosine added MRS broth. Similarly, Lactobacillus sp. 4C10 and Leuconostoc sp. 4E9 did not show arginine decarboxylase activity; however, they produced agmatine in arginine added MRS broth at a level of 338.0 ± 75.3 and 152.3 ± 43.7 mg L–1, respectively. The tyramine amounts produced in the MRS broth supplemented with tyrosine are generally higher than those reported by Lorencová et al.[Citation29] These researchers stated that most of the LAB strains from dairy products produced tyramine lower than 100 mg L–1 in the MRS broth enriched with tyrosine amino acid at the same concentration (0.2%) used in current study, while only Lactobacillus brevis strains isolated from beer produced higher amounts (37–3084 mg L–1) of tyramine.
Table 2. Putrescine, agmatine and tyramine amounts produced by LAB isolates in MRS broth.
Biogenic amine production or decarboxylase activity of bacteria could be determined using differential medium containing pH indicator. The conversion of the color to purple depending on increasing pH is evaluated as decarboxylase positive.[Citation30] Three isolates produced related biogenic amine, although they gave arginine or tyrosine negative results in amino acid decarboxylase test ( and ). Bover-Cid and Holzapfel[Citation31] determined three tyramine producing lactobacilli strains that gave negative results in decarboxylase test. Similar results have been reported by De Llano et al.[Citation32] Such false-negative results may be obtained when the strain produces low amount of amine that is not enough to cause the pH shift for color change as mentioned by Bover-Cid and Holzapfel.[Citation31]
All the LAB isolates produced tyramine in tyrosine added MRS broth. Tyramine formation in foods is associated especially with LAB.[Citation31–Citation33] Also, tyramine has been mentioned as the main biogenic amine produced by LAB strains isolated from different foods, including fermented pork sausage,[Citation28] anchovy,[Citation34] and a fermented cereal-based product “tarhana.”[Citation35] Of the 56 LAB isolates, only one was identified as Leuconostoc sp. 4E9, which was tyrosine decarboxylase positive and produced 158.9 ± 23.1 mg L–1 tyramine in tyrosine added MRS broth ( and ). Leuconostoc species have been previously reported as a tyramine producer.[Citation10,Citation32,Citation36] Moreno-Arribas et al.[Citation10] determined tyramine production (0.8–1.1 g L–1) by Leuconostoc mesenteroides strains in the medium contained corresponding precursor amino acid. In contrast, Lorencová et al.[Citation29] have reported that the L. mesenteroides strain they tested did not produce tyramine. The Streptococcus sp. 4E7 strain was also tyramine producer (). Although there is limited data on biogenic amine production by Streptococcus species, Buňková et al.[Citation37] determined that one of the 21 Streptococcus thermophilus strains produced tyramine.
Putrescine can be formed from ornithine by ornithine decarboxylase or from arginine via arginine decarboxylase.[Citation38] Putrescine formation from arginine can be occurred by two different metabolic pathways. In one of these pathways, arginine is converted to citrulline via arginine deiminase, and then citrulline is converted to ornithine by ornithine transcarbamylase. Finally, putrescine is formed by ornithine decarboxylase from ornithine. In the other pathway, first, agmatine is produced by arginine decarboxylase from arginine. Then, agmatine is either directly converted to putrescine via agmatine deiminase or converted to N-carbamoylputrescine via N-carbamolyputrescine, and then putrescine is formed by N-carbamoylputrescine hydrolase.[Citation8,Citation38] In the current study, although none of the LAB isolates decarboxylated ornithine (), almost all isolates, except Lactobacillus sp. 0A8, produced putrescine at levels from 2.1 ± 0.6 to 33.3 ± 1.7 mg L–1 () in arginine added MRS broth. Therefore, it can be said that these LAB strains produced putrescine from agmatine using the second way that was previously mentioned. In a previous study by Arena et al.,[Citation39] putrescine formation from agmatine by Lactobacillus hilgardii X1B via this pathway has been determined. Likewise, in a very recent study by Ladero et al.,[Citation40] it was reported that all the putrescine producing LAB strains, including Lactobacillus and Lactococcus species, used the agmatine deiminase pathway rather than ornithine decarboxylase pathway.
Conclusions
Results revealed that Lactobacillus species were dominant during shalgam fermentation. The LAB included in shalgam fermentation could be responsible mainly tyramine and agmatine formation. They may contribute to putrescine formation and produced putrescine from agmatine. Additionally, the method for determination the amino acid decarboxylase activity rely on determining the color changes depending on pH changes could give false-negative results. Therefore, the quantitative methods in which the decarboxylation products are detected by HPLC may provide more reliable data.
Funding
This research was supported by Sakarya University Commission for Scientific Research Projects (Project Number 2011-50-01-061).
Additional information
Funding
References
- Erten, H.; Tangüler, H.; Canbaş, A. A Traditional Turkish Lactic Acid Fermented Beverage: Shalgam (Şalgam). Food Review International 2008, 24, 352–359.
- Altay, F.; Karbancioglu-Güler, F.; Daskaya-Dikmen, C.; Heperkan, D. A Review on Traditional Turkish Fermented Non-Alcoholic Beverages: Microbiota, Fermentation Process, and Quality Characteristics. International Journal of Food Microbiology 2013, 167, 44–56.
- Tamang, J.P. Plant-Based Fermented Foods and Beverages of Asia. In Handbook of Plant-Based Fermented Food and Beverage Technology, Hui, Y.H.; Evranuz, Ö.; Eds.; CRC Press Taylor & Francis Group, Boca Raton, FL, 2012; 49–90.
- Erginkaya, Z.; Hammes, W.P. A research on the identification of isolated lactic acid bacteria and on the developing microorganisms during the fermentation of şalgam juice. Gıda 1992, 17, 311–314.
- Tangüler, H.; Erten, H. Occurrence and Growth of Lactic Acid Bacteria Species During the Fermentation of Shalgam (Salgam), a Traditional Turkish Fermented Beverage. LWT–Food Science and Technology 2012, 46, 36–41.
- Fernández, M.; Zúňiga, M. Amino Acid Catabolic Pathways of Lactic Acid Bacteria. Critical Reviews in Microbiology 2006, 32, 155–183.
- Tammam, J.D.; Williams, A.G.; Noble, J.; Lloyd, D. Amino Acid Fermentation in Non-Starter Lactobacillus spp. Isolated From Cheddar Cheese. Letters in Applied Microbiology 2000, 30, 370–374.
- Arena, M.E.; de Nadra, M.C.M. Biogenic Amine Production by Lactobacillus. Journal of Applied Microbiology 2001, 90, 158–162.
- Williams, A.G.; Noble, J.; Banks, J.M. Catabolism of Amino Acids by Lactic Acid Bacteria Isolated from Cheddar Cheese. International Dairy Journal 2001, 11, 203–215.
- Moreno-Arribas, M.V.; Polo, M.C.; Jorganes, F.; Muňoz, R. Screening of Biogenic Amine Production by Lactic Acid Bacteria Isolated from Grape Must and Wine. International Journal of Food Microbiology 2003, 84, 117–123.
- Liu, S.Q.; Holland, R.; Crow, V.L. The Potential of Dairy Lactic Acid Bacteria to Metabolise Amino Acids Via Non-Transaminating Reactions and Endogenous Transamination. International Journal of Food Microbiology 2003, 86, 257–269.
- Landate, J.M.; Pardo, I.; Ferrer, S. Tyramine and Phenylethylamine Production Among Lactic Acid Bacteria Isolated from Wine. International Journal of Food Microbiology 2007, 115, 364–368.
- Silla-Santos, M.H. Biogenic Amines: Their Importance in Foods. International Journal of Food Microbiology 1996, 29, 213–231.
- Ayhan, K.; Durlu-Özkaya, F. Biogenic Amines in Foods. In Metabolism and Applications of Lactic Acid Bacteria, Özer, B.; Ed.; Research Signpost Publishing: Kerala, India, 2008; 87–114.
- Sohrabvandi, S.; Mortazavian, A.M.; Rezaei, K. Health-Related Aspects of Beer: A Review. International Journal of Food Properties 2012, 15, 350–373.
- Özdestan, Ö.; Üren, A. Biogenic Amine Content of Shalgam (Şalgam): A Traditional Lactic Acid Fermented Turkish Beverage. Journal of Agricultural Food Chemistry 2010, 58, 2602–2608.
- İncedayi, B.; Uylaşer, V.; Çopur, Ö.U. A Traditional Turkish Beverage Shalgam: Manufacturing Technique and Nutritional Value. Journal of Food Agriculture and Environment 2008, 6, 31–34.
- Schillinger, U.; Lücke, F.K. Identification of Lactobacilli from Meat and Meat Products. Food Microbiology 1987, 4, 199–208.
- Maijala, R.L. Formation of Histamine and Tyramine by Some Lactic Acid Bacteria in MRS-Broth and Modified Decarboxylation Agar. Letters in Applied Microbiology 1993, 17, 40–43.
- Joosten, H.M.L.J.; Northolt, M.D. Detection, Growth, and Amine-Producing Capacity of Lactobacilli in Cheese. Applied and Environmental Microbiology 1989, 55, 2356–2359.
- Eerola, S.; Hinkkanen, R.; Lindfors, E.; Hirvi, T. Liquid Chromatographic Determination of Biogenic Amines in Dry Sausages. Journal of AOAC International 1993, 76, 575–577.
- Tanguler, H.; Sarıs, P.E.; Erten, H. Microbial, Chemical, and Sensory Properties of Shalgams Made Using Different Production Methods. Journal of the Science of Food and Agriculture 2015, 95, 1008–1015.
- Satomi, M.; Kimura, B.; Mizoi, M.; Sato, T.; Fujii, T. Tetragenococcus Muriaticus sp. Nov., A New Moderately Halophilic Lactic Acid Bacterium Isolated from Fermented Squid Liver Sauce. International Journal of Systematic Bacteriology 1987, 47, 832–836.
- Lonvaud-Funel, A.; Joyeux, A. Histamine Production by Wine Lactic Acid Bacteria: Isolation of a Histamine-Producing Strain of Leuconostoc Oenos. Journal of Applied Microbiology 1994, 77, 401–407.
- Lucas, P.; Wolken, W.A.M.; Claisse, O.; Lolkema, J.S.; Lonvaud-Funel, A. Histamine-Producing Pathway Encoded on An Unstable Plasmid in Lactobacillus Hilgardii 0006. Applied and Environmental Microbiology 2005, 71, 1417–1424.
- Bover-Cid, S.; Miguélez-Arrizado, M.J.; Becker, B.; Holzapfel, W.H.; Vidal-Carou, M.C. Amino Acid Decarboxylation by Lactobacillus Curvatus CTC273 Affected by the pH and Glucose Availability. Food Microbiology 2008, 25, 269–277.
- Romano, A.; Trip, H.; Lolkema, J.S.; Lucas, P.M. Three-Component Lysine/Ornithine Decarboxylation System in Lactobacillus Saerimneri 30a. Journal of Bacteriology 2013, 195, 1249–1254.
- Bover-Cid, S.; Hugas, M.; Izquierdo-Pulido, M.; Vidal-Carau, M.C. Amino Acid-Decarboxylase Activity of Bacteria Isolated from Fermented Pork Sausages. International Journal of Food Microbiology 2001, 66, 185–189.
- Lorencová, E.; Bunkova, L.; Matoulkova, D.; Drab, V.; Pleva, P.; Kuban, V.; Bunka, F. Production of Biogenic Amines by LAB and Bifidobacteria Isolated from Dairy Products and Beer. International Journal of Food Science and Technology 2012, 47, 2086–2091.
- Garai, G.; Dueňas, M.T.; Irastorza, A.; Moreno-Arribas, M.V. Biogenic Amine Production by Lactic Acid Bacteria Isolated form Cider. Letters in Applied Microbiology 2007, 45, 473–478.
- Bover-Cid, S.; Holzapfel, W.H. Improved Screening Procedure for Biogenic Amine Production by Lactic Acid Bacteria. International Journal of Food Microbiology 1999, 53, 33–41.
- De Llano, D.G.; Cuesta, P.; Rodriguez, A. Biogenic Amine Production by Wild Lactococcal and Leuconostoc Strains. Letters in Applied Microbiology 1998, 26, 270–274.
- Leisner, J.J.; Millan, J.C.; Huss, H.H.; Larsen, L.M. Production of Histamine and Tyramine by Lactic Acid Bacteria Isolated form Vacuum-Packed Sugar-Salted Fish. Journal of Applied Microbiology 1994, 76, 417–423.
- Pons-Sánchez-Cascado, S.; Bover-Cid, S.; Veciana-Nogués, M.T.; Vidal-Carau, M.C. Amino Acid-Decarboxylase Activity of Bacteria Isolated Form Ice-Preserved Anchovies. European Food Research and Technology 2005, 220, 312–315.
- Özdestan, Ö.; Üren, A. Biogenic Amine Content of Tarhana: A Traditional Fermented Food. International Journal of Food Properties 2013, 16, 416–428.
- Choudhury, N.; Hansen, W.; Engesser, D.; Hammes, W.P.; Holzapfel, W.H. Formation of Histamine and Tyramine by Lactic Acid Bacteria in Decarboxylase Assay Medium. Letters in Applied Microbiology 1990, 11, 278–281.
- Buňková, L.; Buňka, F.; Hlobilová, M.; Vaňátková, Z.; Nováková, D.; Dráb, V. Tyramine Production of Technological Important Strains of Lactobacillus, Lactococcus, and Streptococcus. European Food Research and Technology 2009, 229, 533–538.
- Landate, J.M.; Arena, M.E.; Pardo, I.; Manca de Nadra, M.C.; Ferrer, S. The Role of Two Families of Bacterial Enzymes in Putrescine Synthesis from Agmatine Via Agmatinase Deiminase. International Microbiology 2010, 13, 169–177.
- Arena, M.E.; Landate, J.M.; Manca de Nadra, M.C.; Pardo, I.; Ferrer, S. Factors Affecting the Production of Putrescine from Agmatine by Lactobacillus Hilgardii X1B Isolated from Wine. Journal of Applied Microbiology 2008, 105, 158–165.
- Ladero, V.; Martin, M.C.; Redruello, B.; Mayo, B.; Florez, A.B.; Fernandez, M.; Alvarez, M.A. Genetic and Functional Analysis of Biogenic Amine Production Capacity Among Starter and Non-Starter LAB Isolated from Artisanal Cheeses. European Food Research and Technology 2015, 241, 377–383.