ABSTRACT
Stingless bees harbor lactic acid bacteria (LAB) which possesses multiple beneficial properties. However, there is no report on LAB in stingless bee from Sabah and their products. This study aimed to isolate LAB from several stingless bee species and their products as well as to perform characterization and safety assessments. A total of 104 strains were isolated and seven potential antimicrobial LAB isolates were identified from stingless bee and their products. Characterization, identification, and assessments were performed on seven (A2b, B3b, P1b, H4b, A6, B5, and B10) LAB that exert potential antimicrobial activity against Listeria monocytogenes ATCC 7644. The A6 isolate was closely related to the Lactobacillus pentosus species whereas isolates B5 and B10 were closely related to Weissella paramesenteroides species. Finally, isolates P1b, H4b, B3b, and A2b were closely related to the Enterococcus sp. These seven LAB isolates were able to survive in stimulated gastrointestinal tract conditions (acidic, salt, bile salt, and temperature). The carbon fermentation, proteolytic activities, acidification, milk coagulation, and wide antibiotic susceptibility testing of seven LAB isolates revealed their potential used as a probiotic and fermentation purposes. Therefore, more studies are warranted to investigate the potential of these LAB isolates towards applications in probiotic and fermentation.
Introduction
Lactic acid bacteria (LAB) are generally present in many fermented and non-fermented food products.[Citation1] Thus, these microorganisms benefits the food industry and also carry health benefits towards humans such as enhancement of the immune system, improves digestion, have anticancer activities and antidiabetic effects.[Citation2–5] Therefore, LAB are a group of bacteria and a suitable choice to be used in food application as well as a probiotic.
LAB are characterized as catalase negative and strictly fermentative bacteria. They produce a mixture of lactic acid, carbon dioxide, acetic acid, and ethanol (hetero-fermentation) or produce lactic acid only (homo-fermentation) as their major metabolic end-products.[Citation6,Citation7] When LAB were consumed, they would inhabit the intestinal mucosal membrane and it can also be found in the reproductive tracts of humans and animals, providing various benefits to the host organism.[Citation1,Citation7] As part of the commensal bacteria, LAB affect the health of the gastrointestinal tract, especially the small and large intestines, by enhancing the hydrolysis of lactose to glucose and galactose that favour rapid absorption and fermentation by the bacteria in the gut microflora.[Citation8] Moreover, in this study, Escherichia coli (E. coli)ATCC 11775, Listeria monocytogenes (L. monocytogenes) ATCC 7644, and Salmonella enterica (S. enterica) ATCC 13076 were used as indicator strains to identify the potential antimicrobial activity of the LAB strains. S. enterica and E. coli are Gram-negative whereas L. monocytogenes is Gram-positive. They are commonly found as food-borne pathogens.[Citation9–11]
Stingless bees are a tropical rainforest pollinator distributed in both tropical and subtropical regions.[Citation12] There are approximately 35 stingless bee species in Peninsular Malaysia and roughly 32 species of stingless bees in Borneo.[Citation13,Citation14] The most dominant stingless bee species are Heterotrigona itama followed by Geniotrigona thoracica, Tetragonula laeviceps, Lepidotrigona terminata, and Tetrigona apicalis. Stingless bees are widely domesticated for the meliponiculture industry to harvest stingless bees' products, namely honey, bee bread, and propolis.[Citation14,Citation15] Stingless bees have been reported to harbor up to 50 novel LAB species belonging to the genera of Lactobacillus and Bifidobacterium.[Citation16] These LAB have potential benefits in various applications. However, the characteristics of LAB from stingless bees in North Borneo, Sabah sre unknown. Therefore, in this study we isolated, characterized, and assessed LAB isolated from the stingless bees in Sabah.
Materials and methods
Sample collection and isolation of lactic acid bacteria
Three species of stingless bees (Geniotrigona thoracica, Heterotrigona itama, and Tetrigona binghami) with their bee products namely bee bread, honey, bee propolis were collected. The stingless bees were located from two areas, an urban area and a deep tropical forest situated in Kiulu, Kota Kinabalu, Sabah (North Borneo). Bee eggs were collected from Heterotrigona itama inhabiting at urban area only. The stingless bees were washed with 95% (v/v) ethanol for surface sterilization and the gut was dissected and homogenized. Approximately, 0.1 g of each bee's bread, honey, eggs and propolis was weighed and homogenized.
LAB were isolated as described previously with slight modifications.[Citation17] Briefly, homogenized samples were diluted by 10-fold and inoculated in De man, Rogosa and Sharpe (MRS) and M17 broths (Merck, Darmstadt, Germany) under anaerobic conditions at 30°C for 48 hrs. After incubation, the broth culture was serially diluted from 10−2 until 10−6 using 0.9% (w/v) of sterile saline water. Roughly, 100 µL of the diluents were inoculated on MRS and M17 agar supplemented with 0.01% (w/v) of sodium azide as a suppression reagent followed by anaerobic incubation at 30°C for 48 hrs. A single colony was picked and sub-cultured as a pure isolate.
Screening of lactic acid bacteria
A total of 104 bacterial strains were isolated from three species of stingless bees and their products (T. binghami, N = 48; H. itama, N = 35; G. thoracica, N = 21) (Tables S1–S3). Screening of LAB were performed using Gram-staining, morphology observations and catalase test. Gram-staining and the catalase test were performed as described by Hasali et al.[Citation17] Briefly, a single colony culture from an agar plate was fixed on a glass slide and stained with crystal violet for 60 s. The slide was washed with distilled water and stained with iodine for 60 s followed by decolourization with absolute ethanol. Then the slide was then flooded with safranin for 45 s and washed with distilled water. The slide was viewed under a light microscope. The catalase test was performed by placing a single colony culture on a glass slide followed by drop of 3% (v/v) hydrogen peroxide was dropped on the culture. The slide was observed for the formation of bubbles which indicated a positive catalase reaction or vice versa. The in vitro tests and safety assessments were designed and conducted according to a FAO/WHO report.[Citation18]
Antimicrobial activity assessment
The antimicrobial activity of isolated strains (N = 104) was performed using agar well diffusion assay in triplicate as described previously.[Citation17] E. coli ATCC 11775, L. monocytogenes ATCC 7644, and S. enterica ATCC 13076 were used as indicator bacteria. A pre-prepared solidified Brain Heart Infusion (BHI) (Merck, Darmstadt, Germany) agar was poured with a layer of 6 mL BHI agar which was inoculated with 60 µL of indicator bacteria with 0.5 McFarland. Next, a 6 mm hole was made in the agar. Isolated LAB were cultured in MRS broth overnight. Cell-free culture supernatants were prepared by centrifugation of the overnight cultures at 12,000 × g for 20 min at 4°C.[Citation19] The supernatant (100 µL) was seeded into the hole and incubated at 37°C for 24 hrs. The zone of growth inhibition was measured and recorded.
Molecular identification of lactic acid bacteria
The genus or species of seven isolated LAB (A6, B5, B10, P1b, H4b, B3b and A2b) were identified using polymerase chain reaction (PCR) targeting the 16S rRNA gene. Briefly, gDNA was isolated according to Vingataramin & Frost.[Citation20] Approximately, 100 µL of broth culture were mixed with 450 µL of extraction solution (240 mM NaOH, 2.7 mM EDTA, 74% Ethanol) in a 1.5 mL microcentrifuge tube followed by heating at 80°C for 10 min. The mixture was centrifuged at 16,060 × g for 10 min and decanted. The pellet of each sample was resuspended with an optimized suspension solution containing 0.1 mM EDTA, 50 mM Tris-HCl, pH 8.0, 1% Triton-X-100, and 0.5% Tween-20.
The resuspended pellet was used as a template for PCR amplification of the 16S rRNA gene using a forward primer (27 F: 5ʹ-AGAGTTTGATCCTGGCTCAG-3ʹ) and a reverse primer (1492 R: 5ʹ-GGTTACCTTGTTACGACTT-3ʹ) as described previously.[Citation21] Briefly, a 25 µL total reaction volume containing 0.3 pmol of forward and reverse primers, 400 µM of dNTPs, 0.5 U of Taq polymerase (Invitrogen Inc., USA), 1X PCR buffer, 1 µL of gDNA template, and 1.5 mM of MgCl2 were prepared. The PCR conditions were as follows: initial denaturation of one cycle at 94°C for 2 min, 30 cycles of 95°C for 10 s; 53°C for 30 s; 72°C for 1 min, final extension of 1 cycle at 72°C for 5 min. The PCR products were purified and sequenced using BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems, USA). The sequences were compared and aligned using MUSCLE in MEGA X version 10.5.1 software. Gene sequences from NCBI GeneBank DNA database were obtained and phylogenetic tree was constructed using maximum-likelihood method with 1000 times bootstraps.
Biochemical characterization of lactic acid bacteria
Carbon fermentation test, starch hydrolysis, proteolytic activity and milk acidification assay were performed on seven isolated LAB (A6, B5, B10, P1b, H4b, B3b and A2b) in triplicates as previously described.[Citation22]
Carbon fermentation test
Nutrient agar supplemented with 1% (w/v) of glucose and 0.004% of bromocresol purple was prepared. Approximately, 10 µL of LAB overnight culture was placed on the agar and incubated overnight at 37°C. The appearance of yellow colouration around the culture indicates consumption of the carbon source.
Starch hydrolysis
Isolated LAB and control strains were streaked on nutrient agar supplemented with 2% (w/v) soluble starch powder and incubated overnight at 37°C. A small amount of iodine was placed on the culture and formation of a clear zone around the culture indicated a positive reaction.
Proteolytic activity
Proteolytic activity of LAB was determined by inoculation of culture onto skim milk agar and incubated at 37°C overnight. Clear zones around the culture indicated the presence of proteolytic activity.
Milk acidification
Skim milk broth culture of the LAB was prepared and incubated at 37°C up to 72 hrs. Skim milk without addition of LAB was used as a contol. Parameters such as aroma, color, coagulation, and pH of the culture were observed and measured every 24 h.
Screening for probiotic properties
Seven isolates (A6, B5, B10, P1b, H4b, B3b and A2b) were screened for probiotic properties. Acid resistance, salt tolerance, and temperature sensitivity of LAB were conducted in triplicates as described previously with modifications.[Citation23] Bile salt tolerance test was performed in triplicates as described by Pavli et al.[Citation24]
Acid tolerance
Approximately 1 mL of LAB culture was inoculated in MRS broth and anaerobically incubated at 37°C overnight. The overnight culture (~1.7 × 1010 until ~8.6 × 1010 CFU/mL) was sub-cultured into MRS broth with different pH levels (2, 3, and 4) and incubated overnight at 37°C. After 24 hrs of incubation, 10 µL of the culture was grown on MRS agar using the track plate technique and incubated at 37°C for 48 hrs. The number of viable cell counts (CFU/mL) were determined.
Salt tolerance
LAB were inoculated into 10 mL of MRS broth culture containing different salt (NaCl) concentrations, from 0% until 12% and supplemented with 0.004% bromocresol purple as an coloured indicator and incubated at 37°C for 48 hrs. The growth of LAB was inspected visually for color changes. The salt tolerance level of the LAB isolated were evaluated via absorbance reading at the optical density (OD) of 600nm.
Temperature sensitivity
Isolated LAB were cultured in 10 mL MRS broth containing 0.004% bromocresol purple at different temperatures (4°C, 25°C, 28°C, 37°C, and 80°C) for 24 hrs. LAB growth was observed for color change. The temperature sensitivity of LAB was determined by recording absorbance readings OD 600nm.
Bile acid tolerance
Bile salt tolerance was conducted by streaking fresh culture of isolated LAB on MRS agar supplemented with 0.3% (w/v) of taurodeoxycholic acid (bile acid) and incubate at aerobic and anaerobic conditions at 37°C for 24 hrs. The test was performed in triplicates.
Safety assessments
Antibiotic susceptibility
Antibiotic susceptibility of seven isolated LAB (A6, B5, B10, P1b, H4b, B3b and A2b) was performed in triplicates as described by Angmo et al.[Citation25] Approximately 100 µL of LAB culture containing 1 × 108 CFU/mL were spread on M17 agar plate in triplicates. Twenty-four kinds of antibiotic discs (amounts) as listed in the following were placed on the agar plate and incubated at 37°C for 24 hrs; penicillin G (2 units), erythromycin (10 µg), vancomycin (30 µg), ampicillin (25 µg), ceftriaxone (30 µg), colistin sulfate (10 µg), streptomycin (10 µg), amikacin (30 µg), norfloxacin (10 µg), chloramphenicol (30 µg), tetracycline (10 µg), nalidixic acid (30 µg), gentamycin (30 µg), mecilinam (25 µg), nitrofurantoin (300 µg), sulfamethoxazole/trimethoprim (25 µg), kanamycin (30 µg), neomycin (30 µg), lincomycin (10 µg), cloxacillin (5 µg), ciprofloxacin (10 µg), cefuroxime sodium (30 µg), bacitracin (10 µg), and novobiocin (30 µg). The zones of inhibition were measured using a caliper and categorized as sensitive (S, ≥21 mm), intermediate (I, 16–20 mm), or resistant (R, ≤15 mm) as described by Liasi et al.[Citation19] and the Clinical and Laboratory Standards Institute (CLSI).[Citation26]
Blood hemolysis
The ability of isolated LAB to lyse blood cells were conducted in triplicate as described by a previous study with slight modifications.[Citation24] Seven isolated LAB (A6, B5, B10, P1b, H4b, B3b and A2b) cultures were streaked onto Columbia agar plates containing 5% (w/v) of horse blood in triplicates and incubated at 37°C for 24 hrs. The agar was observed for signs of alpha (α), beta (β), and gamma (γ) hemolytic.
Results and discussion
Isolation of lactic acid bacteria
Both Gram-positive and Gram-negative bacteria were isolated from T. binghami, H. itama, G. thoracica bees and their products. Microscopic observation of the 104 bacterial isolates found 18 Gram-negative and 86 Gram-positive bacteria (). All bacteria isolated from H. itama were Gram-positive. This is in line with previous studies, where only Gram-positive isolates were found in H. itama.[Citation17,Citation27] There were 29 coccus and 75 rod-shaped bacteria among the isolates. Supported by previous studies which also isolated LAB from Apis mellifera L. bees, the bacterial isolates were either coccus or rod shaped.[Citation28,Citation29] The catalase test from this study showed that 12 and 92 bacterial isolates were catalase positive and negative, respectively. One of the interesting characteristics of LAB is their anaerobic properties which can be tested using the catalase test. The 92 catalase negative of our LAB indicated that the isolates were anaerobic. Catalase negative LAB have demonstrate the potential to exert antimicrobial activity and used in food applications.[Citation17,Citation27,Citation30]
Table 1. Summary of bacterial strains isolated from stingless bee in urban and deep tropical forest area
Antimicrobial assay against pathogenic bacteria
Another objective of this study was to identify LAB that exhibit antimicrobial activity against pathogenic bacteria. The antagonistic activity of 104 bacterial isolates was tested against three pathogenic bacteria (E. coli ATCC 11775, L. monocytogenes ATCC 7644, and S. enterica ATCC 13076). Interestingly, regardless of the stingless bee species, we found that the seven isolates (A6, B5, B10, P1b, H4b, B3b and A2b) exhibited antimicrobial activities against the Gram-positive Li. monocytogenes ATCC 7644 with inhibition zones (mm ± S.D.) ranging from 7.50 ± 0.02 to 9.80 ± 0.28 (). These LAB isolates were from bees which inhabited deep tropical forests and of various sources such as gut, bee bread, propolis, and honey. This finding was in line with a past study which reported that LAB isolates that showed inhibition activity against Staphylococcus epidermdiis, Pseudomonas aeruginosa, and L. monocytogenes.[Citation17] On the contrary, we observed that all bacterial isolates (N = 104) did not exhibit antimicrobial activity against Gram-negative E. coli ATCC 11775 and S. enterica ATCC 13076. This Gram-specific activity was highly interesting. However, further investigations are needed to identify the exact mode of actions and the chemical compounds involved in the inhibition, which is of the limitations of this study. One could only suspect that bacteriocin may have played a role in the antibacterial activity demonstrated by the isolates, due to its narrow bioactivity spectrum which only act on either Gram-positive or Gram-negative bacteria.[Citation31,Citation32] However, there are several instances where a bacteriocin could inhibit both Gram-positive and Gram-negative pathogens.[Citation33]
Table 2. Antimicrobial activity of the isolated bacterial strains against Listeria monocytogenes ATCC 7644
Molecular identification of isolates using 16S rRNA gene sequence analysis
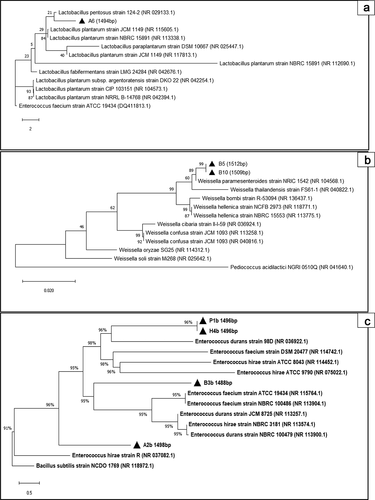
We performed molecular identification of the seven isolates (A6, B5, B10, P1b, H4b, B3b and A2b) which exhibited antimicrobial properties against pathogenic bacteria. BLAST and phylogenetic tree analysis identified isolate A6 to be closely related to the Lactobacillus pentosus species (). Isolate B5 and B10 were closely related to Weissella paramesenteroides species (). Isolates P1b, H4b, B3b, and A2b were closely related to the Enterococcus sp. (). These identified species are widely reported as lactic acid bacteria (LAB). Praet et al.[Citation34] also isolated Lactobacillus sp. and Weisella sp. from the bee gut. Enterococcus sp. was also isolated from H. itama in another previous study.[Citation35] Supporting our results, Lactobacillus sp., Weisella sp., and Enterococcus sp. have been demonstrated to produce novel bacteriocins which exhibit antimicrobial activities against a wide range of pathogens as well as antibiotic-resistant bacteria such as vancomycin-resistant enterococci.[Citation36–38]
Figure 1. Phylogenetic tree of LAB isolates and their related taxa based on 16S rRNA sequence. The phylogenetic trees were constructed using the Maximum-likelihood method (MEGA X 10.1.5). Numbers in the parentheses indicates the accession number of published sequences. (A) Phylogenetic tree of A6 isolate. (B) Phylogenetic tree of B5 and B10 isolates. (C) Phylogenetic tree of P1b, H4b, B3b, A2b isolates
Biochemical characterization of isolated lactic acid bacteria
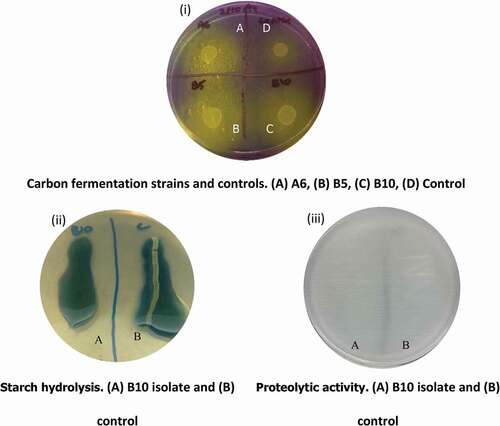
The carbon fermentation test found that all isolated LAB were able to utilize glucose as their carbon source ( (i)). However, we observed that all isolated LAB were not able to hydrolyze starch and did not contain proteolytic activity as indicated by the absence of clear zones ( (ii & iii)). These results suggested that the isolated LAB in our study utilize glucose as their main carbon source and did not contain amylase to facilitate starch hydrolysis as well as proteolytic activity. A previous study also reported that all of the studied LAB isolates were able to consume monosaccharide (glucose) and only 27% of their isolates were able to breakdown polysaccharide, such as starch.[Citation22] Furthermore, honey mainly contains disaccharides and monosaccharides which created a positive selective pressure for the LAB to selectively contain enzymes for the degradation of monosaccharide and disaccharide degradation.[Citation39] Hence, polysaccharide (starch) degrading activities were not observed in our LAB isolates. In addition, LAB have also been found to contain amylase has been purified for food applications.[Citation40] The proteolytic activity of LAB is one of the many ways for the bacteria to acquire amino acids for growth, survival and produce of amino acid supplements.[Citation30,Citation41] The absence of proteolytic activity in our isolated LAB was due to the carbohydrate-rich and not protein-rich environment of samples from stingless bees and their products. . This was demonstrated by a previous study which successfully isolated 61.3% of LAB containing proteolytic activity from dairy products, which have opposite environment as those in bees and their products.[Citation42]
Figure 2. Various test using isolated LAB and control strain (L. monocytogenes ATCC 7644). An example of (i) carbon fermentation test where the change of color in yellow indicates carbon consumption, (ii) starch hydrolysis test where the clear zone in (B) indicates a positive reaction, (iii) proteolytic activity test which clear zone indicates the presence of proteolytic activity
For LAB to be applied in the dairy industry, the acidification and milk coagulation process is important. We have shown that the overall acidifying effect of all seven strains was ranged from the initial pH value of approximately 6.4 to final pH value of roughly 4.1 (). The acidification of milk offers a microorganism and pathogen control which causes spoilage of fermented products.[Citation30] The authors also reported that the pH of milk fermentation did not reach below pH 4.0 which was in line with our observations. Therefore, our LAB isolates (A2b, B3b, P1b, H4b, A6, and B10) with rapid acidification within 24 hrs are good candidates for food production.
Table 3. Milk acidification activity of seven LAB strains
Milk coagulation time for the isolated LAB ranged from 24 hrs to 72 hrs with isolates A2b, B3b, P1b, H4b and A6 which comprised of Lactobacillus sp. and Enterococcus sp. as the fastest coagulators. Several Lactobacillus sp., such as Lactobacillus helveticus, Lactobacillus fermentum, Lactobacillus plantarum, and Lactobacillus mesenteroides have been reported as fast acid producing and milk coagulation strains.[Citation43] The acidification and milk coagulation generates various aroma, which were from various kinds of bacterial metabolites, such as acetone, acetoin, acetic acid, and propanoic acids.[Citation44]
Probiotic properties of isolated lactic acid bacteria
Acid and bile salt tolerance
The tolerance levels of LAB in gastric-like acidic and bile salt-rich environments are crucial properties for probiotic cultures in order to exert its beneficial effects in the intestine because such conditions are stressful for survival.[Citation45] As shown in , the survivability of the isolated LAB indicated poor to high tolerance toward acidic environments. A6, B5, and B10 LAB isolates have a high tolerance in acidic environments where there was no substantial differences in the viable cell count at acidic conditions (pH at 2, 3, and 4) when compared to their respective control. The current findings supported that the LAB isolates have high survival in the acidic conditions of the human gut (pH 2 to 5) as the transit time in food along the human gut is a maximum of 3 hrs.[Citation46] However, A2b, B3b, P1b and H4b LAB isolates have a poor tolerance toward low pH level which were not ideal probiotics as shown by a low viability cell count of ~2.0 × 109 CFU/mL when compared to their respective controls with a viability of ~2.0 × 1010 CFU/mL ().
Table 4. Number of viable cell count (CFU/mL) (± S.D.) at different acidity levels
Intestinal bile salts have been reported to inhibit bacterial growth by disruption of the cellular membrane. We found that the seven LAB isolates were able to hydrolyze bile acid without the change of morphology in aerobic and anaerobic conditions, suggesting a tolerance toward bile salts. Franz et al.[Citation47] reported the presence of bile salt tolerance in Enterococcus strains which were also found in our study. Bacteria that are able to survive in bile salts can utilize bile salts for metabolic activities to support the growth and colonization of the intestinal tract which will elicit beneficial effects to the host.[Citation48]
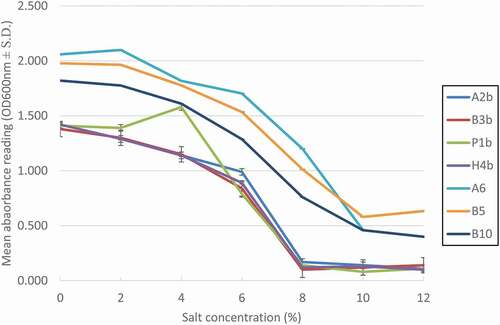
Salt tolerance
The intestinal tract does not contain only bile salts but also contains high levels of salt (NaCl).[Citation23] The seven isolated strains have a salt tolerance level of up to 6% as shown by the bacterial growth (). The growth of the isolated strains has a steady growth decline and suffers from significantly reduced growth at 8% (A2b, B3b, P1b, H4b) and 10% (A6, B5, B10) of NaCl. Our findings are in line with a previous study where they reported their bacterial isolates were able to thrive at a salt concentration of 4–6%. However, there are LAB isolates that are highly sensitive to salt concentrations, whereby they could not grow in salt concentration higher than 2%.[Citation49] Salt tolerance in LAB is an important factor for food applications. High salt concentration culture could cause loss of turgor pressure in cells which affects the enzyme activity, water activity, and physiology.[Citation50] Therefore, a high salt tolerance level of LAB is crucial in commercial purposes. During fermentation processes, lactic acid is produced by LAB, and thus alkaline solution would be added into the culture broth to prevent acidic conditions. Here, the free acid would be converted to its salt form, thereby increasing the osmotic pressure of the cell.[Citation51]
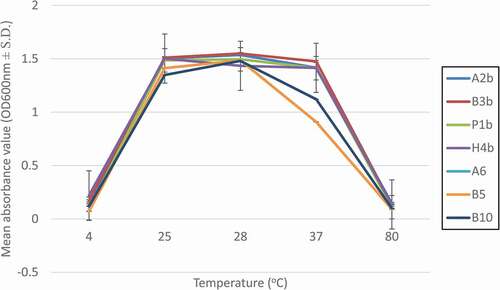
Temperature tolerance
Tolerance to temperature is another important factor to evaluate in LAB isolates that have to survive in the human gastrointestinal tract or survive throughout the industrial food processes. illustrates the growth of the seven isolated LAB at different temperatures. A2b, B3b, P1b, H4b, and A6 LAB isolates have optimal growth from 25°C to 37°C. Whereas, LAB isolates B5 and B10 have an optimal growth from 25°C to 28°C and suboptimal growth at 37°C. We observed that a low temperature (4°C) and an extreme high temperature (80°C) were unfavorable to sustain the growth of all seven LAB isolates indicated that they were not psychrophile or thermophile. Supported by previous authors, they reported that LAB isolates are able to survive within the range of 25°C to 40°C and could not survive under extreme temperatures. The isolates from that study were being used as poultry probiotics.[Citation52]
Safety assessment
The antibiotic susceptibility of all seven LAB isolates towards 24 antibiotics were determined for safety purposes. The antibiotics were categorized into five groups by their mode of actions. The groups were cell wall inhibitors, protein synthesis inhibitors (30s and 50s subunits), DNA synthesis inhibitors, folic acid synthesis inhibitors, and multiple mechanisms. Antimicrobial resistance strains raise due to gene transmission of transposons, plasmids, and bacterial gene mutations.[Citation53] We observed that the antibiotic susceptibility profile was correlated with the source of stingless bee species. H. itama that harbours A2b, B3b, P1b, and H4b LAB isolates have the same susceptibility profile toward all antibiotics with different mode of actions (). A6 and B10 LAB were isolated from G. thoracica and B5 LAB was isolated from T. binghami. These three isolates had different antibiotic susceptibility profiles when compared with each other as well as with the LAB isolates from H. itama. Antibiotic resistant LAB strains are not recommended for food fermentation as they could serve as a reservoir for antibiotic resistant genes.[Citation54] This reservoir of antibiotic resistant genes could cause the widespread of multi-antibiotic resistance bacteria by genetic transferring to nonresistant bacteria.[Citation55,Citation56] However, the usage of antibiotic resistant LAB as a probiotic bacterium is beneficial because bacteria can survive during antibiotic treatment which could act as an adjuvant treatment to exert its beneficial effects during the course of antibiotic treatment for eradication of Helicobacter pylori.[Citation57] Additionally, all seven LAB isolates possess γ-hemolytic activity on blood agar plate, which are considered safe to be consumed ().[Citation24] Non-hemolytic LAB have been used in fermenting of coconut palm nectar as well as meat products.[Citation24,Citation58] Therefore, the antimicrobial resistance profile and hemolytic activity of these seven LAB isolates provided an initial insight toward choosing the right LAB isolate for future applications. However, the one limitation of the study are data regarding the presence of antibiotic resistance gene was not assessed.
Table 5. Antibiotic susceptibility and hemolytic activity of LAB isolates
Conclusion
This study had isolated, identified, andcharacterized seven lactic acid bacteria (LAB) from stingless bees and their products. Supported by the experiments in our study, these LAB isolates have great potential to be utilized as a probiotic source for humans, animals use as well as in starter cultures in food applications in the future.
Supplemental information
Supplementary information has been provided as follows; Table S1-Bacteria isolated from Tetrigona binghami collected in urban and deep forest area, Table S2-Bacteria strains isolated from Heterotrigona itama collected in urban and deep tropical forest area, Table S3-Isolated bacteria from Geniotrigona thoracica collected in deep forest.
Supplemental Material
Download MS Word (29.5 KB)Acknowledgments
This study was supported by Universiti Malaysia Sabah internal grants (SBK0384-2018 & SDK0134-2020). Lucky Poh Wah Goh: data curation, writing-original draft, writing-review & editing; Arnold Marshall Molujin: investigation, data curation, visualization; Kaliswaran Muthu: investigation, data curation, visualization; Rahmath Abdulla: conceptualization, writing-review & editing; Mohd Khalizan Sabullah: conceptualization, writing-review & editing; Ainol Azifa Mohd. Faik: conceptualization, writing-review & editing; Jualang Azlan Gansau: conceptualization, writing-review & editing; Roslina Jawan: conceptualization, supervision, writing-review & editing. The authors declare that they have no conflict of interest.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Mojgani, N.; Hussaini, F.; Vaseji, N. Characterization of Indigenous Lactobacillus Strains for Probiotic Properties. Jundishapur J. Microbiol. 2015, 8, e17523. DOI: https://doi.org/10.5812/jjm.17523.
- Jager, R.; Purpura, M.; Farmer, S.; Cash, H. A.; Keller, D. Probiotic Bacillus coagulans GBI-30, 6086 Improves Protein Absorption and Utilization. Probiotics Antimicrob. Proteins. 2018, 10, 611–615. DOI: https://doi.org/10.1007/s12602-017-9354-y.
- Kamiya, T.; Watanbe, Y.; Makino, S.; Kano, H.; Tsuji, N. M. Improvement of Intestinal Immune Cell Function by Lactic Acid Bacteria for Dairy Products. Microorganisms. 2017, 5, 1. DOI: https://doi.org/10.3390/microorganisms5010001.
- Lakritz, J. R.; Poutahidis, T.; Levkovich, T.; Varian, B. J.; Ibrahim, Y. M.; Chatzigiagkos, A.; Mirabal, S.; Alm, E. J.; Erdman, S. E. Beneficial Bacteria Stimulate Host Immune Cells to Counteract Dietary and Genetic Predisposition to Mammary Cancer in Mice. Int. J. Cancer. 2014, 135, 529–540. DOI: https://doi.org/10.1002/ijc.28702.
- Niibo, M.; Shirouchi, B.; Umegatani, M.; Morita, Y.; Ogawa, A.; Sakai, F.; Kadooka, Y.; Sato, M. Probiotic Lactobacillus gasseri SBT2055 Improves Insulin Secretion in a Diabetic Rat Model. J. Dairy Sci. 2019, 102, 997–1006. DOI: https://doi.org/10.3168/jds.2018-15203.
- Ali, A. A.;. Beneficial Role of Lactic Acid Bacteria in Food Preservation and Human Health: A Review. Res. J. Microbiol. 2010, 5, 1213–1221. DOI: https://doi.org/10.3923/jm.2010.1213.1221.
- Narvhus, J. A.; Axelsson, L. Lactic Acid Bacteria. In Encyclopedia of Food Sciences and Nutrition; Caballero, B., Ed.; Academic Press: USA, 2003, pp 3465–3472. DOI: https://doi.org/10.1016/B0-12-227055-X/00673-8.
- Flint, H. J.; Scott, K. P.; Duncan, S. H.; Louis, P.; Forano, E. Microbial Degradation of Complex Carbohydrates in the Gut. Gut Microbes. 2012, 3, 289–306. DOI: https://doi.org/10.4161/gmic.19897.
- Luna-Guevara, J. J.; Arenas-Hernandez, M. M. P.; Martínez De La Peña, C.; Silva, J. L.; Luna-Guevara, M. L. The Role of Pathogenic E. Coli in Fresh Vegetables: Behavior, Contamination Factors, and Preventive Measures. Int. J. Microbiol. 2019, 1–10. DOI: https://doi.org/10.1155/2019/2894328.
- Matle, I.; Mbatha, K. R.; Madoroba, E. A. Review of Listeria monocytogenes from Meat and Meat Products: Epidemiology, Virulence Factors, Antimicrobial Resistance and Diagnosis. Onderstepoort. J. Vet. Res. 2020, 87, 1869. DOI: https://doi.org/10.4102/ojvr.v87i1.1869.
- Ford, L.; Moffatt, C. R. M.; Fearnley, E.; Miller, M.; Gregory, J.; Sloan-Gardner, T. S.; Polkinghorne, B. G.; Bell, R.; Franklin, N.; Williamson, D. A.; et al. The Epidemiology of Salmonella enterica Outbreaks in Australia, 2001–2016. Front. Sustain. Food Syst. 2018, 2, 86. DOI: https://doi.org/10.3389/fsufs.2018.00086.
- Eltz, T.; Brühl, C. A.; Imiyabir, Z.; Linsenmair, K. E. Nesting and Nest Trees of Stingless Bees (Apidae: meliponini) in Lowland Dipterocarp Forests in Sabah, Malaysia, with Implications for Forest Management. Forest Ecol. Manag. 2003, 172, 301–313. DOI:https://doi.org/10.1016/S0378-1127(01)00792-7.
- Schwarz, H. F. 1937. Results of the Oxford University Sarawak (Borneo) Expedition: Bornean Stingless. Bull. Am. Mus. Nat. Hist. LXXIII. 281–329.
- Jaapar, M. F.; Halim, M.; Mispan, M. R.; Jajuli, R.; Saranum, M. M.; Zainuddin, M. Y.; Ghazi, R.; Ghani, I. A. The Diversity and Abundance of Stingless Bee (Hymenoptera: meliponini) in Peninsular Malaysia. Adv. Environ. Biol. 2016, 10, 1–7.
- Kelly, N.; Farisya, M. S.; Kumara, T. K.; Marcela, P. Species Diversity and External Nest Characteristics of Stingless Bees in Meliponiculture. Pertanika J. Trop. Agric. Sci. 2014, 37, 293–298.
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R. J.; Flaberg, E.; Szekely, L.; Olofsson, T. C. Symbionts as Major Modulators of Insect Health: Lactic Acid Bacteria and Honeybees. PLoS ONE. 2012, 7, e33188. DOI: https://doi.org/10.1371/journal.pone.0033188.
- Hasali, N. M.; Zamri, A. I.; Lani, M. N.; Mubarak, A.; Suhaili, Z. Identification of Lactic Acid Bacteria from Meliponine Honey and Their Antimicrobial Activity against Pathogenic Bacteria. Am.-Eurasian J. Sustain. Agri. 2015, 9, 1–6.
- FAO/WHO. Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation. 2001, 1–51.
- Liasi, S. A.; Azmi, T. I.; Hassan, M. D.; Shuhaimi, M.; Rosfarizan, M.; Ariff, A. B. Antimicrobial Activity and Antibiotic Sensitivity of Three Isolates of Lactic Acid Bacteria from Fermented Fish Product, Budu. Malaysian J. Microbiol. 2009, 5, 33–37.
- Vingataramin, L.; Frost, E. H. A Single Protocol for Extraction of gDNA from Bacteria and Yeast. BioTechniques. 2015, 58, 120–125. DOI: https://doi.org/10.2144/000114263.
- Reysenbach, A.-L.; Ehringer, M.; Hershberger, K. Microbial Diversity at 83oC in Calcite Springs, Yellowstone National Park: Another Environment Where the Aquificales and “Korarchaeota” Coexist. Extremophiles. 2000, 4, 31–67.
- Abbasiliasi, S.; Tan, J. S.; Ibrahim, T. A.; Ramanan, R. N.; Vakhshiteh, F.; Mustafa, S.; Ling, T. C.; Rahim, R. A.; Ariff, A. B. Isolation of Pediococcus acidilactici Kp10 with Ability to Secrete Bacteriocin-like Inhibitory Substance from Milk Products for Applications in Food Industry. BMC Micro. 2012, 12, 260. DOI: https://doi.org/10.1186/1471-2180-12-260.
- Ayo-Omogie, H.; Okorie, E. In Vitro Probiotic Potential of Autochthonous Lactic Acid Bacteria and Microbiology of Kunu Made from Mixed Grains. Br. Microbiol. Res. J. 2016, 14, 1–10. DOI: https://doi.org/10.9734/bmrj/2016/25403.
- Pavli, F. G.; Argyri, A. A.; Papadopoulou, O. S. Probiotic Potential of Lactic Acid Bacteria from Traditional Fermented Dairy and Meat Products: Assessment by in Vitro Tests and Molecular Characterization. J. Prob. Health. 2016, 4, 3. DOI: https://doi.org/10.4172/2329-8901.1000157.
- Angmo, K.; Kumari, A.; Savitri,; Bhalla, T. C. Probiotic Characterization of Lactic Acid Bacteria Isolated from Fermented Foods and Beverage of Ladakh. LWT Food Sci. Technol. 2016, 66, 428–435. DOI: https://doi.org/10.1016/j.lwt.2015.10.057.
- CLSI. 2016. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100S. 26th. Wayne, PA: Clinical and Laboratory Standards Institute, pp 1-249.
- Aween, M. M.; Hassan, Z.; Muhialdin, B. J.; Eljamel, Y. A.; Al-Mabrok, A. S. W.; Lani, M. N. Antibacterial Activity of Lactobacillus acidophilus Strains Isolated from Honey Marketed in Malaysia against Selected Multiple Antibiotic Resistant (MAR) Gram-Positive Bacteria. J. Food Sci. 2012, 77, M364–M371. DOI: https://doi.org/10.1111/j.1750-3841.2012.02776.x.
- Carina Audisio, M.; Torres, M. J.; Sabaté, D. C.; Ibarguren, C.; Apella, M. C. Properties of Different Lactic Acid Bacteria Isolated from Apis mellifera L. Bee-gut. Microbiol. Res. 2011, 166, 1–13. DOI: https://doi.org/10.1016/j.micres.2010.01.003.
- Kenfack, C.; Kaktcham, P.; Ngoufack, F.; Wang, Y.; Yin, L.; Zhu, T. Screening and Characterization of Putative Probiotic Lactobacillus Strains from Honey Bee Gut (Apis mellifera). J. Adv. Microbiol. 2018, 10, 1–18. DOI: https://doi.org/10.9734/jamb/2018/40780.
- Jawan, R.; Kasimin, M. E.; Jalal, S. N.; Mohd Faik, A. A.; Abbasiliasi, S.; Ariff, A., Isolation, Characterisation and in Vitro Evaluation of Bacteriocins-producing Lactic Acid Bacteria from Fermented Products of Northern Borneo for Their Beneficial Roles in Food Industry, J. Phys. Conf. Ser., 2019, IOP. 1358, 012020.
- Zacharof, M. P.; Lovitt, R. W. Bacteriocins Produced by Lactic Acid Bacteria a Review Article. APCBEE Procedia. 2012, 2, 50–56. DOI: https://doi.org/10.1016/j.apcbee.2012.06.010.
- Perez, R. H.; Zendo, T.; Sonomoto, K. Novel Bacteriocins from Lactic Acid Bacteria (LAB): Various Structures and Applications. Microb. Cell Fact. 2014, 13, S3. DOI: https://doi.org/10.1186/1475-2859-13-s1-s3.
- Vieco-Saiz, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and Inputs from Lactic Acid Bacteria and Their Bacteriocins as Alternatives to Antibiotic Growth Promoters during Food-Animal Production. Front. Microbiol. 2019, 10, 57. DOI: https://doi.org/10.3389/fmicb.2019.00057.
- Praet, J.; Meeus, I.; Cnockaert, M.; Houf, K.; Smagghe, G.; Vandamme, P. Novel Lactic Acid Bacteria Isolated from the Bumble Bee Gut: Convivina Intestini Gen. Nov., Sp. Nov., Lactobacillus bombicola sp. Nov. And Weissella bombi sp. Nov. Antonie Van Leeuwenhoek. 2015, 107, 1337–1349. DOI: https://doi.org/10.1007/s10482-015-0429-z.
- Mohammad, S. M.; Mahmud-Ab-Rashid, N.-K.; Zawawi, N. Probiotic Properties of Bacteria Isolated from Bee Bread of Stingless Bee Heterotrigona itama. J. Apic. Res. 2021, 60, 172–187. DOI: https://doi.org/10.1080/00218839.2020.1801152.
- Gaspar, C.; Donders, G. G.; Palmeira-de-oliveira, R.; Queiroz, J. A.; Tomaz, C.; Martinez-de-oliveira, J.; Palmeira-de-oliveira, A. Bacteriocin Production of the Probiotic Lactobacillus acidophilus KS400. AMB Express. 2018, 8, 153. DOI: https://doi.org/10.1186/s13568-018-0679-z.
- Srionnual, S.; Yanagida, F.; Lin, L.-H.; Hsiao, K.-N.; Chen, Y. Weissellicin 110, a Newly Discovered Bacteriocin from Weissella cibaria 110, Isolated from Plaa-Som, a Fermented Fish Product from Thailand. Appl. Environ. Microbiol. 2007, 73, 2247–2250. DOI: https://doi.org/10.1128/aem.02484-06.
- Phumisantiphong, U.; Siripanichgon, K.; Reamtong, O.; Diraphat, P. A Novel Bacteriocin from Enterococcus faecalis 478 Exhibits A Potent Activity against Vancomycin-resistant Enterococci. PLoS One. 2017, 12, e0186415. DOI: https://doi.org/10.1371/journal.pone.0186415.
- Shamsudin, S.; Selamat, J.; Sanny, M.; Abd. Razak, S.-B.; Jambari, N. N.; Mian, Z.; Khatib, A. Influence of Origins and Bee Species on Physicochemical, Antioxidant Properties and Botanical Discrimination of Stingless Bee Honey. Int. J. Food Prop. 2019, 22, 239–264. DOI: https://doi.org/10.1080/10942912.2019.1576730.
- Padmavathi, T.; Bhargavi, R.; Priyanka, P. R.; Niranjan, N. R.; Pavitra, P. V. Screening of Potential Probiotic Lactic Acid Bacteria and Production of Amylase and Its Partial Purification. J. Genet. Eng. Biotechnol. 2018, 16, 357–362. DOI: https://doi.org/10.1016/j.jgeb.2018.03.005.
- Lim, Y. H.; Foo, H. L.; Loh, T. C.; Mohamad, R.; Abdullah, N. Comparative Studies of Versatile Extracellular Proteolytic Activities of Lactic Acid Bacteria and Their Potential for Extracellular Amino Acid Productions as Feed Supplements. J. Animal Sci. Biotechnol. 2019, 10, 15. DOI: https://doi.org/10.1186/s40104-019-0323-z.
- García-Cano, I.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Wang, K.; Kosmerl, E.; Jiménez-Flores, R. Lactic Acid Bacteria Isolated from Dairy Products as Potential Producers of Lipolytic, Proteolytic and Antibacterial Proteins. Appl. Microbiol. Biotechnol. 2019, 103, 5243–5257. DOI: https://doi.org/10.1007/s00253-019-09844-6.
- Akabanda, F.; Owusu-Kwarteng, J.; Tano-Debrah, K.; Parkouda, C.; Jespersen, L. The Use of Lactic Acid Bacteria Starter Culture in the Production of Nunu, a Spontaneously Fermented Milk Product in Ghana. Int. J. Food Sci. 2014, 2014, 1–11. DOI: https://doi.org/10.1155/2014/721067.
- Routray, W.; Mishra, H. N. Scientific and Technical Aspects of Yogurt Aroma and Taste: A Review. Compr. Rev. Food Sci. Food F. 2011, 10, 208–220. DOI: https://doi.org/10.1111/j.1541-4337.2011.00151.x.
- Jena, P. K.; Trivedi, D.; Thakore, K.; Chaudhary, H.; Giri, S. S.; Seshadri, S. Isolation and Characterization of Probiotic Properties of Lactobacilli Isolated from Rat Fecal Microbiota. Microbiol. Immunol. 2013, 57, 407–416. DOI: https://doi.org/10.1111/1348-0421.12054.
- Begum, S. B.; Roobia, R. R.; Karthikeyan, M.; Murugappan, R. M. Validation of Nutraceutical Properties of Honey and Probiotic Potential of Its Innate Microflora. LWT Food Sci. Technol. 2015, 60, 743–750. DOI: https://doi.org/10.1016/j.lwt.2014.10.024.
- Franz, C. M. A. P.; Specht, I.; Haberer, P.; Holzapfel, W. H. Bile Salt Hydrolase Activity of Enterococci Isolated from Food: Screening and Quantitative Determination. J. Food Prot. 2001, 64, 725–729. DOI: https://doi.org/10.4315/0362-028x-64.5.725.
- Shehata, M. G.; El Sohaimy, S. A.; El-Sahn, M. A.; Youssef, M. M. Screening of Isolated Potential Probiotic Lactic Acid Bacteria for Cholesterol Lowering Property and Bile Salt Hydrolase Activity. Ann. Agric. Sci. 2016, 61, 65–75. DOI: https://doi.org/10.1016/j.aoas.2016.03.001.
- Samedi, L.; Charles, A. L. Isolation and Characterization of Potential Probiotic Lactobacilli from Leaves of Food Plants for Possible Additives in Pellet Feeding. Ann. Agric. Sci. 2019, 64, 55–62. DOI: https://doi.org/10.1016/j.aoas.2019.05.004.
- Coulibaly, I.; Robin, D. D.; Destain, J.; Philippe, T. Characterization of Lactic Acid Bacteria Isolated from Poultry Farms in Senegal. Afr. J. Biotechnol. 2008, 7, 2006–2012. DOI: https://doi.org/10.5897/ajb2008.000-5048.
- Mohd Adnan, A. F.; Tan, I. K. P. Isolation of Lactic Acid Bacteria from Malaysian Foods and Assessment of the Isolates for Industrial Potential. Bioresour. Technol. 2007, 98, 1380–1385. DOI: https://doi.org/10.1016/j.biortech.2006.05.034.
- Reuben, R. C.; Roy, P. C.; Sarkar, S. L.; Alam, R.-U.; Jahid, I. K. Isolation, Characterization, and Assessment of Lactic Acid Bacteria toward Their Selection as Poultry Probiotics. BMC Microbiol. 2019, 19, 253. DOI: https://doi.org/10.1186/s12866-019-1626-0.
- Teuber, M.; Meile, L.; Schwarz, F. Acquired Antibiotic Resistance in Lactic Acid Bacteria from Food. Vol. 76. Netherlands, Antonie Van Leeuwenhoek. 1999, 115–137. doi:https://doi.org/10.1023/a:1002035622988.
- Wang, K.; Zhang, H.; Feng, J.; Ma, L.; Fuente-Núñez, C.; De La, Wang, S.; Lu, X. Antibiotic Resistance of Lactic Acid Bacteria Isolated from Dairy Products in Tianjin, China. J. Agric. Food Res. 2019, 1, 100006. DOI: https://doi.org/10.1016/j.jafr.2019.100006.
- Bujnakova, D.; Strakova, E.; Kmet, V. In Vitro Evaluation of the Safety and Probiotic Properties of Lactobacilli Isolated from Chicken and Calves. Anaerobe. 2014, 29, 118–127. DOI: https://doi.org/10.1016/j.anaerobe.2013.10.009.
- Mathur, S.; Singh, R. Antibiotic Resistance in Food Lactic Acid Bacteria—a Review. Int. J. Food Microbiol. 2005, 105, 281–295. DOI: https://doi.org/10.1016/j.ijfoodmicro.2005.03.008.
- Du, Y.-Q.;. Adjuvant Probiotics Improve the Eradication Effect of Triple Therapy For Helicobacter pylori infection. World J. Gastroenterol. 2012, 18, 6302. DOI: https://doi.org/10.3748/wjg.v18.i43.6302.
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B. V.; Sreenivasa, M. Y. Probiotic Properties of Lactic Acid Bacteria Isolated from Neera: A Naturally Fermenting Coconut Palm Nectar. Front. Microbiol. 2019, 10, 1382. DOI: https://doi.org/10.3389/fmicb.2019.01382.