Abstract
Xylophagous termites are capable of degrading lignocellulose by symbiotic gut microorganisms along with the host’s indigenous enzymes. Therefore, the termite gut might be a potential niche to obtain natural yeasts with celluloytic, xylanolytic and ethanologenic traits required for bioethanol production from lignocellulosic biomass. In this study, we cultured 79 yeasts from three different termites viz. Coptotermes heimi, Odontotermes javanicus and Odontotermes obesus. After suitable screening methods, we identified 53 yeasts, which belonged to 10 genera and 16 different species of both ascomycetous and basidiomycetous yeasts. Most yeasts in the present study represent their first-ever isolation from the termite gut. Representative strains of identified yeasts were evaluated for their cellulolytic, xylanolytic, and ethanologenic abilities. None of the isolates showed cellulase activity; 22 showed xylanolytic activity, while six produced substantial quantities of ethanol. Among xylanolytic cultures, Pseudozyma hubeiensis STAG 1.7 and Hannaella pagnoccae STAG 1.14 produced 1.31 and 1.17 IU of xylanase. Among ethanologenic yeasts, the strains belonging to genera Candida and Kodamaea produced high amount of ethanol. Overall, highest ethanol level of 4.42 g/L was produced by Candida tropicalis TS32 using 1% glucose, which increased up to 22.92 g/L at 35 °C, pH 4.5 with 5% glucose. Fermentation of rice straw hydrolysate gave 8.95 g/l of ethanol with a yield of 0.42 g/g using the strain TS32. Our study highlights the gut of wood-feeding termites as a potential source of diverse yeasts that would be useful in the production of xylanase and bioethanol.
Keywords:
1. Introduction
Yeasts populate many different habitats on our planet. Some yeast species can be found in high numbers in different habitats, while others may be restricted to only a few specialized areas or niches, suggesting that the overall yeast distribution is not uniform [Citation1]. The yeast diversity of unique or rare habitats has not been studied as much as that of common habitats likes soil, plants, other terrestrial and aquatic habitats [Citation2]. The gut of insects, especially wood-feeders (xylophagous), is one such niche for yeasts that has not been fully realized in terms of its diversity and biotechnological potential. In the past two decades, researchers have delved into the gut microbiome of wood-feeding insects like beetles and wood roaches and have successfully isolated a large number of yeasts, including many novel species [Citation3–5]. The insect–yeast association is of a mutualistic type wherein the yeasts assist the insect host in many metabolic pathways, provide nutrients, detoxify allelochemicals, and promote their expansion into nutrient-poor substrates. Insects also help the yeasts in various aspects like protection against biotic stress, dispersal of spores, and facilitating outbreeding of yeasts. Many ascomycetous yeasts have been known to be associated with the gut of insects [Citation6].
The gut of wood-feeding termites mimics a bioreactor and represents a niche that harbors a plethora of microorganisms like bacteria, protozoa, and fungi. These symbiotic microbes produce multiple hydrolytic enzymes to digest the cellulosic and hemicellulosic components of wood [Citation7]. These enzyme complexes, along with the endogenous enzymes produced by the termite, effectively carry out the breakdown of lignocellulose into simple monomeric sugars like glucose, xylose, arabinose, galactose, and mannose. The association between some termites and fungi is well known, but only a few reports on yeast–termite associations have been published [Citation8–11]. Early reports show that yeast species belonging to the genera Candida, Debaryomyces, Pichia, and Sporothrix were obtained from the hindgut of lower termites [Citation9]. Recently, novel species Sugiyamaella mastotermitis and Papiliotrema odontotermitis were isolated from the gut of Mastotermes darwiniensis and Odontotermes obesus, respectively [Citation10]. It is speculated that yeast symbionts might also be capable of producing lignocellulolytic enzymes to convert cellulose and hemicellulose into simpler monomeric units.
The production of second-generation ethanol is impeded by the absence of a microorganism capable of producing enzymes for the complete hydrolysis of hemicelluloses and fermenting the different sugars present in lignocellulosic biomass [Citation12]. Since yeast is the most suitable candidate for ethanol production, it is desirable for it to produce lignocellulose degrading enzymes and effectively ferment both hexose and pentose sugars derived from lignocellulosic hydrolysates. One way to achieve this is through genetic engineering of a yeast strain with cellulolytic and xylanolytic activities and employing these recombinant strains for simultaneous saccharification and fermentation to produce bioethanol. An alternative to this is the use of wild type yeasts that can initially produce hydrolytic enzymes, which will efficiently breakdown pretreated lignocellulose, utilize monomeric sugars for growth, and ferment these sugars to produce ethanol. This is a promising strategy; however, to recover such unique yeasts is a challenging task. Screening more wild type yeasts with such desirable traits are the need-of-the-hour. As mentioned earlier, the termite gut might harbor yeasts with such desirable traits. Till now, there has been only one study, which suggests the gut of wood-feeding termite can be a niche for yeasts with desirable traits like xylan degradation and utilization of monomeric sugars for ethanol production [Citation11]. However, there has been no systematic study to assess the cellulolytic, xylanolytic and ethanologenic properties of yeasts from different species of termites. Further, termite gut-associated yeasts have never been tested for their potential to produce ethanol directly from the lignocellulosic hydrolysate. Therefore, the present study aimed to identify, characterize, and assess the biotechnological potential of yeasts harbored in the gut of wood-feeding termites, especially those yeasts that can produce cellulase, xylanases, and ethanol.
2. Materials and methods
2.1. Termite collection and isolation of yeasts
In the present study, wood-feeding termites were collected from rotting wood at Agharkar Research Institute, Pune, Maharashtra (18.5207451°N, 73.8315643°E), and Singalandhapuram, Namakkal District, Tamil Nadu (11.420428°N, 78.220487°E). The identity of the termites was established by DNA sequencing of the mitochondrial COII gene [Citation13] and the sequences were submitted in GenBank. The protocols followed for harvesting the termite gut for isolating yeasts have been previously described [Citation5]. Thirty worker termites from each sample were surface sterilized with 95% ethanol, and their guts were removed aseptically, crushed, and transferred to 0.7% saline. 100 µL of the gut suspension was streaked on yeast extract-peptone-dextrose (YPD; yeast extract 10 g/L, peptone 20 g/L, dextrose 20 g/L, and agar 15 g/L; pH 5.5) containing antibiotics (200 µg/mL streptomycin, 25 µg/mL ampicillin, and 25 µg/mL chloramphenicol) and the plates were incubated at 25 °C for 2–5 days. Single colonies were streaked for purification, and glycerol stocks of the strains were preserved at −80 °C until further use.
2.2. Morphological and biochemical characterization of yeast strains
For micromorphological characterization, cultures were grown on yeast extract-malt agar (YM; 3.0 g/L yeast extract, 3.0 g/L malt extract, 10 g/L glucose, and 15.0 g/L agar; pH 5.0), 5% malt-extract agar (HiMedia Laboratories, Mumbai, India), and corn-meal agar (HiMedia Laboratories), at 26 °C and studied with a differential interference contrast (DIC) microscope (BX53, Olympus, Tokyo, Japan) equipped with a DP73 camera (Olympus) and cellSens 1.13 imaging software. Standard protocols were employed for the biochemical characterization of yeasts [Citation1]. Assimilation tests were performed in duplicate in liquid media (6.7 g/L yeast nitrogen base (YNB) and 11.7 g/L yeast carbon base (YCB)) and results were noted after 7 days of incubation. Seven different sugars (d-glucose, d-galactose, maltose, mannose, sucrose, l-arabinose, and d-xylose) were used to determine the fermentation profiles of the selected yeasts and results were noted after 7 days.
2.3. Identification and phylogenetic analyses of yeasts
The yeasts were characterized according to their cell morphology followed by PCR-RFLP patterns of the ITS-rDNA region using the restriction enzyme MspI (New England Biolabs, Ipswich, MA). The strains with identical RFLP banding patterns were grouped and initially considered to belong to the same species. At least one representative strain from each grouping was selected and subjected to sequence analysis of the partial large subunit (LSU) rRNA gene. Genomic DNA of yeasts was isolated using a standard protocol [Citation14]. PCR amplification of the LSU rRNA gene was carried out using standard primers and PCR cycling conditions [Citation15]. The PCR products were purified using a GenElute PCR Clean-Up Kit (Sigma-Aldrich, St. Louis, MO) and sequenced with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA). Phylogenetic analyses were made with MEGA 7 [Citation16] using the Maximum Likelihood algorithm and the Tamura–Nei evolutionary model, as suggested by the implemented model test.
2.4. Determination of xylanase and cellulase activity
Yeasts under study were evaluated for the production of xylanase and cellulase following previously described methods with slight modifications [Citation17,Citation18]. Yeast cells were inoculated (at OD600 nm = 0.5) in YNB-xylan (YNB 6.7 g/L and xylan 10 g/L; pH 5.5) and YNB-carboxymethyl cellulose (YNB 6.7 g/L and CMC 10 g/L; pH 5.5) media to induce xylanase and CMCase production at 30 °C, respectively. After 72 h, the culture supernatant was obtained by centrifugation (7000 x g for 15 min) and used to determine the enzyme activities. The assay mixture consisted of 200 µL of culture supernatant containing crude enzyme and 1800 µL of xylan (5 g/L) suspension in 100 mM sodium phosphate buffer (pH 7.0). Cellulase assay was performed similarly, with 500 µL of culture supernatant and 1500 µL of CMC (5 g/L) suspension in 100 mM sodium phosphate buffer. The mixtures were incubated at 45 °C for 30 min, followed by immediate chilling on ice to stop the reaction. The amount of reducing sugars released was determined using the 3, 5-dinitrosalicylic acid method [Citation19]. One unit of enzyme activity was defined as the amount of enzyme required to release 1 μmol reducing sugars per min under the assay conditions. All enzymatic measurements were performed in triplicate.
2.5. Optimization of ethanol production
The ethanol-producing potential of the yeasts was tested by following previously described methods with slight modifications [Citation20]. Experiments were carried out in 120 mL serum bottles with a working volume of 30 mL fermentation media, which consisted of 5 g/L yeast extract, 1 g/L peptone, 2 g/L (NH4)2SO4, 5 g/L KH2PO4, 0.2 g/L MgSO4·7H2O, and 10 g/L glucose. The pH of the medium was adjusted to 5.0 using 2 N HCl. Yeast cells were grown in yeast extract peptone glucose medium (7 g/L yeast extract, 14 g/L peptone, and 10 g/L glucose), and inoculated (at OD600 nm = 0.2) into fermentation media. These bottles were incubated at 30 °C on a rotary shaker at 150 rpm. Ethanol concentration in fermentation media was determined after 72 h of incubation by gas chromatography (450-GC gas chromatograph, Bruker, Billerica, MA). The flame ionization detector (FID) was maintained at temperatures of 150 °C and 200 °C. The oven temperature was maintained at 80 °C. Ethanol (Sigma-Aldrich) was used as the internal standard. All experiments were performed in triplicates. Important parameters like ethanol yield (Yp/s, g/g), ethanol productivity (Qp, g/L/h), and fermentation efficiency (%) for all strains were also calculated [Citation21]. To compare ethanol yield of the selected strain, a standard reference strain Saccharomyces cerevisiae NCIM3186 was also used. The quantitative effects of temperature, pH, and reducing sugar concentration were evaluated for the highest ethanol-producing strain to find an optimum condition for ethanol production. Ethanol production at 20 °C, 25 °C, 30 °C, 35 °C, 37 °C, and 40 °C were analyzed. After optimization of the fermentation temperature, the optimum pH of fermentation media for maximum ethanol yield was determined by analyzing ethanol production at pH 4.0, 4.5, 5.0, 5.5, and 6.0. Finally, the optimum concentration of sugar (glucose) was determined by using varying concentrations of glucose (5%, 10%, 15%, 20%, and 30%) in the fermentation broth and assessing their effects on ethanol yield. Data were analyzed by one-way Analysis of Variance (ANOVA) with GraphPad Prism 5 Statistics Software. Differences with a p value <0.05 were considered statistically significant.
2.6. Ethanol production from lignocellulosic biomass
Lignocellulosic hydrolysates (LH) were prepared by following a previously described method, with slight modifications [Citation22]. Pretreatment of rice straw was carried out with alkali, followed by enzymatic saccharification using commercially available enzyme cocktail. Rice straw (RS) biomass was mixed with 1% NaOH at biomass loading of 10.0% w/v and autoclaved at 121 °C for 45 min. The slurry obtained was filtered using muslin or cheesecloth; biomass residues were washed with sterile distilled water until neutral pH was achieved and then dried thoroughly before further processing. For saccharification, 1.5 g of the dried pretreated RS biomass was mixed with 30 mL Na-citrate buffer (50 mM, pH 4.8) using 15 FPU Cellulase Blend (Sigma-Aldrich) per gram of dried pretreated RS biomass in 120 mL serum bottles. The saccharification was performed at 50 °C for 72 h with continuous shaking at 150 rpm. Before fermentation, the saccharified slurry was supplemented with 5 g/L KH2PO4, 0.12 g/L MgSO4, 2.0 g/L (NH4)2SO4, 1 g/L mycological peptone and 5 g/L yeast extract. The highest ethanol-producing strain was selected for this experiment. Overnight grown yeast cells (O.D600 =1.00) of the selected strain was used as inoculum. Fermentation was carried out at 35 °C for 72 h with shaking at 150 rpm, and the production of ethanol was analyzed by using GC. The released sugars before and after fermentation were estimated using an HPLC-UPLC system (Shimadzu, Kyoto, Japan) equipped with a Refractive Index (RI) detector. The column used was Aminex HPX-87H (300 × 7.8 mm) (Bio-Rad Laboratories, Hercules, CA) kept at a temperature of 40 °C, and the mobile phase was 5 mM H2SO4 at a flow rate of 0.7 mL/min. Data were processed and analyzed using real-time software (LabSolutions CS, Shimadzu, Kyoto, Japan). The concentrations of sugars were analyzed against standard calibration curves using standards of Supelco/Sigma, USA.
3. Results and discussion
3.1. Termite collection and identification
The BLASTn analysis of the mitochondrial COII gene sequences of three different termites identified them as O. obesus (MN913606), O. javanicus (MN913608) and C. heimi (MN913607) respectively. O. obesus is one of the most widely found termite species in South Asia. Despite being generally known as fungus-growing insects, termites of the genus Odontotermes are also destructive wood-boring pests in the tropics and subtropics of Africa and Asia, especially the Indian subcontinent. The species C. heimi belong to the family Rhinotermitidae, which are popular wood-feeding lower termites [Citation23].
3.2. Identification and phylogenetic analyses of yeasts
We isolated a total of 79 yeasts, which included 19 from O. obesus, 35 from O. javanicus and 25 from C. heimi. Screening by PCR-RFLP of ITS-rDNA region resulted in 53 genotypes out of 79 strains, wherein the strains with identical RFLP banding patterns were grouped together as one genotype. Therefore, 53 yeasts; sixteen from O. obesus, 22 from O. javanicus and fifteen from C. heimi, were selected as representative strains and further identified by DNA sequencing of partial LSU rRNA gene (). The DNA sequencing results showed that the 53 yeasts belonged to 10 genera and 16 different species. Since there were multiple strains of 16 different yeast species, we short-listed 23 strains representing all sixteen yeast species for further biotechnological studies (). The identities of the shortlisted strains were confirmed by a maximum likelihood-based phylogenetic analysis, which correctly placed all the 23 yeasts under Ascomycota and Basidiomycota phyla (). Candida tropicalis was the most frequently occurring yeast to be isolated from C. heimi. The other yeast species obtained from this termite were Kodamaea ohmeri, T. coremiiforme, and T. asahii. Three different species of yeasts were isolated from O. obesus; Sugiyamaella smithiae, S. valenteae, and P. manshurica. Maximum yeast diversity was obtained from O. javanicus, where yeasts from genus Papiliotrema were the most frequently occurring ones, represented by three different species; P. flavescens, P. mangalensis, and P. laurentii. Other yeasts isolated from O. javanicus were Vishniacozyma taibaiensis, Pseudozyma hubeiensis, H. luteola, H. pagnoccae, V. dimennae, and Cystobasidium slooffiae. Interestingly, yeasts obtained from O. obesus were ascomycetous, while those isolated from O. javanicus belonged to the Basidiomycota group. This is noteworthy because both termites belonged to the same genus and were sampled from the same geographical location. This suggests that the yeast flora in the gut of termites may depend on the host species and not on the geographical location of the host. However, it would be imperative to determine the yeast flora of a large number of different termite species from varied geographical areas so as to give a more conclusive insight to understand the ecology of termite gut-associated yeasts. Initial reports indicated that ascomycetous yeasts like Candida, Sporothrix, Debaryomyces, and Pichia were obtained from termites belonging to the families Termopsidae, Mastotermitidae, Hodotermitidae, Kalotermitidae and Rhinotermitidae [Citation8,Citation9]. More recently, eighteen yeast species, including seven novel species belonging to the genera Candida, Hamamotoa, Meyerozyma, and Sugiyamaella, were reported from the termite R. chinensis [Citation11]. Our study is the first report of yeasts being isolated from the termite C. heimi and O. javanicus. Yeasts from the genera Vishniacozyma, Kodamea, Pseudozyma, Hannaella, and Cystobasidium have been reported for the first time from termites through this study. It is interesting to note that majority of yeasts isolated from the three termites belonged to the classes Tremellomycetes and Saccharomycetes. Yeasts from the Sugiyamaella and Papiliotrema genera have been previously isolated from Mastotermes darwiniensis and O. obesus, respectively [Citation10], however, this is the first report of S. valenteae, P. flavescens, P. mangalensis, and P. laurentii being isolated from termites.
Figure 1. Phylogenetic placement of yeasts under study based on partial LSU-rRNA gene sequences. The tree was reconstructed using the maximum-likelihood analysis of 647 aligned positions with the Tamura–Nei model. The scale bar indicates the number of expected substitutions per site. The numbers provided on branches are frequencies with which a given branch appeared in 1000 bootstrap replications. The tree was rooted with Rhizopus oryzae.
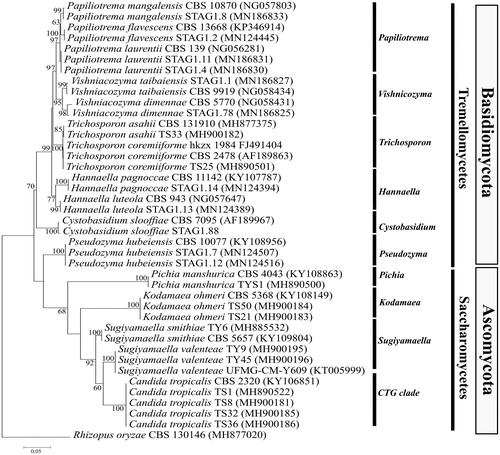
Table 1. Summary of yeasts obtained in this study and their biochemical profile.
3.3. Morphological and biochemical characterization of yeast strains
The yeast strains showed varied morphological characteristics on YM agar plates (Supplementary Figure S1). Other micromorphological characteristics like ascospore formation, hyphae/pseudohyphae formation were observed by culturing on different media (Supplementary Table S1). Several growth tests were performed to compile a comprehensive biochemical profile of yeast strains isolated in this study (Supplementary Table S2). All the tested isolates displayed growth on glucose, xylose, and mannose, except Vishniacozyma dimennae, which did not assimilate mannose (). Only two isolates did not assimilate galactose; T. asahii and Pichia manshurica. Most strains utilized maltose and sucrose. All four strains of C. tropicalis (TS1, TS8, TS32, and TS36) fermented glucose, galactose, and maltose. Glucose and sucrose were fermented by both K. ohmeri strains (TS21 and TS50). Glucose and xylose are the most abundant sugars in the lignocellulosic hydrolysates (LH), while other monosaccharides (arabinose, galactose, and mannose) and disaccharides (cellobiose) are present in trace amounts. Therefore, assimilation and fermentation of all the sugars present in the LH is essential to develop an economically feasible ethanol production strategy from lignocellulosic biomass. Keeping this in mind, we analyzed the carbon source utilization potential of yeast isolates with both pentoses as well as hexoses. We observed that they could efficiently utilize these sugars for their growth, which may help in efficient ethanol production from lignocellulosic biomass.
3.4. Xylanase and cellulase activity
The enzymatic activities of all the short-listed strains have been summarized in . High xylanase activity was exhibited by some strains using YNB-xylan media. Pseudozyma hubeiensis STAG 1.7 and H. pagnoccae STAG 1.14 showed the highest xylanase activities of 1.31 and 1.17 IU respectively at 45 °C. These levels were comparable to the xylanase activity of Candida pseudorhagii (1.73 IU) and C. gotoi (1.13 IU), which were isolated from the gut of wood-feeding termites and reported with xylanase activities higher than that of recombinant yeast strains [Citation11]. Papiliotrema mangalensis STAG 1.8 (0.89 IU) and K. ohmeri TS21 (0.79 IU) also produced significant levels of xylanase. Other yeast strains exhibited little or no xylanase activity under xylan substrate induction. Pseudozyma hubeiensis was previously known to produce high levels of xylanases [Citation24], and this is the first report of it being isolated from the termite gut. Yeasts like P. hubeiensis and H. pagnoccae might be important symbionts in the gut of xylophagous termites which help them hydrolyze xylan to xylose. Endo-1,4-β-xylanase (EC 3.2.1.8) and β-xylosidase (EC 3.2.1.37) are the major xylanases that are known for their extracellular hydrolytic activities against xylan, especially in yeasts and other fungi [Citation25]. It is well-known that the symbiotic flagellates participate in cellulose break down in the termite gut; however, there has been a gap in knowledge about the microbes responsible for xylanolytic processes. Our observations suggest that some yeasts residing in the gut of termites can be responsible for xylan degradation. These xylanolytic yeasts can be potential candidates for industrial-scale production of xylanase and along with their cellulolytic counterparts, can play a significant role in the complete hydrolysis and effective breakdown of the lignocellulosic biomass for sustainable ethanol production. Cellulase (CMCase) activity was absent in all the yeast strains under study. There are very few yeasts known to produce high levels of cellulases [Citation26], and to the best of our knowledge, there are no reports of cellulolytic yeasts isolated from the gut of wood-feeding termites. However, we believe that more extensive and elaborative sampling of gut associated yeasts from different termite species might fetch us yeasts, which can produce both xylanase and cellulase.
Table 2. Ethanol and xylanase production profiles of yeasts.
3.5. Optimization of ethanol production
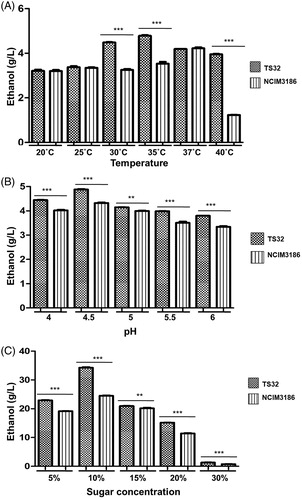
On evaluating the ethanol production of all strains under study using 1% glucose, it was found that six strains belonging to two different species showed high ethanol titers. The fermentation parameters like ethanol concentration, yield, productivity, and fermentation efficiency have been summarized in . The highest ethanol production was achieved at 72 h, and ethanol titers obtained ranged from 3.02 to 4.42 g/L. Candida tropicalis produced the highest levels of ethanol with strains TS32 and TS36, producing 4.42 g/L and 4.31 g/L of ethanol with fermentation efficiencies of 86.50 and 84.34%, respectively. Two more strains of C. tropicalis TS8 and TS1 produced 4.07 and 3.78 g/L of ethanol with fermentation efficiencies of 79.64 and 73.97%, respectively. Kodamea ohmeri strains TS21 and TS50 also produced significant levels of ethanol; 3.97 and 3.02 g/L, respectively. The basidiomycetous yeasts failed to ferment glucose to produce ethanol just as the ascomycetous yeasts in the study failed to show significant xylanase activity. Fermentation of xylose to ethanol by termite gut-associated yeasts has been studied previously [Citation11], but fermentation of glucose has not been assessed before the present study. Based on the fermentation efficiencies of all strains, C. tropicalis TS32 was selected for all further experiments since it produced the highest amount of ethanol. To compare the ethanol yield obtained with each varying parameter, a standard reference strain Saccharomyces cerevisiae NCIM3186 was also used, and the ethanol production of both strains has been depicted in . After evaluating the effect of fermentation parameters related to ethanol production, it was found that the optimum fermentation temperature for the test strain TS32 was 35 °C (), at which it produced 4.78 g/L of ethanol with a fermentation efficiency of 93.54% (Supplementary Table S3). The ethanol productivity decreased drastically at 20 and 40 °C. The optimum pH for fermentation was found to be 4.5 (). At this pH, TS32 produced high level of ethanol, i.e., 4.88 g/L, with a fermentation efficiency as high as 95.50%. When varying concentrations of glucose were used in fermentation media, it was found that the highest yield (0.46 g/g) and ethanol efficiency of 89.71% were obtained with 5% glucose (). Fermentation media supplemented with 10% glucose gave the highest concentration of ethanol (34.29 g/L), but the efficiency decreased to 67%. Ethanol production was inhibited at glucose concentration above 10% (i.e., 15% and 20%), giving low yields and decreased efficiency. Fermentation carried out with 30% glucose gave negligible amount of ethanol for both strains; TS32 and NCIM3186. We observed that ethanol production is drastically reduced at high sugar concentrations. This may be a consequence of osmotic stress exerted on yeast cells at high glucose concentrations. Interestingly, under all test parameters, the ethanol titer produced by C. tropicalis TS32 was higher as compared to the reference standard strain S. cerevisiae NCIM3186.
Figure 2. Effect of temperature, pH, and sugar concentration on ethanol production. (A) Ethanol production (in g/L) by TS32 and standard strain NCIM3186 tested at different temperatures; 20, 25, 30, 35, 37, and 40 °C. (B) Ethanol production (in g/L) at 35°C and different pH tested; 4.0, 4.5, 5.0, 5.5, and 6.0. (C) Ethanol production (in g/L) at 35 °C, pH 4.5 and varying glucose concentrations; 5%, 10%, 15%, 20%, and 30%. Asterisks (*) symbol denotes the level of significance with a p value <0.05 analyzed by one-way ANOVA.
3.6. Ethanol production from lignocellulosic biomass
We also set up experiments to assess ethanol production from lignocellulosic biomass hydrolysate using C. tropicalis TS32, which gave the highest level of ethanol in previous experiments. Being one of the most abundant, renewable forms of lignocellulosic biomass in India, rice straw was used for this study. Rice straw consists of lignin (5–24%), cellulose (32–47%) and hemicellulose (19–27%) [Citation27]. Pretreatment of rice straw was carried out to get rid of lignin and make the cellulosic/hemicellulosic fractions available for hydrolytic action of enzymes. Enzymatic hydrolysis or saccharification of the RS released various monomer sugars which can be fermented. Glucose is the major monomeric sugar produced after hydrolysis of LC biomass and is easily fermented by most yeasts. Other sugars like xylose, arabinose, and sometimes disaccharides like cellobiose are also released, but in lesser proportions as compared to glucose [Citation28]. Thus, the effective fermentation of glucose is necessary for high ethanol yields. During our study, the total sugar content released post pretreatment and enzymatic hydrolysis was 33.94 g/L (20.94 g/L glucose, 10.60 g/L xylose, and 2.40 g/L arabinose) (). Cellobiose was absent in the LH aliquots when tested by HPLC. After 72 h of fermentation of LC biomass, the ethanol titer produced by C. tropicalis TS32 strain was 8.95 g/L with a yield of 0.42 g/g and efficiency of 83.56%, while that of the reference strain S. cerevisiae NCIM3186 was 8.65 g/L with 0.41 g/g yield and 80.76% efficiency (). This is noteworthy since this is the first report of a yeast strain associated with the gut of wood-feeding termite with the ability to produce ethanol directly from LC biomass hydrolysate.
Table 3. Ethanol production from alkali-pretreated rice straw hydrolysate.
4. Conclusion
The metabolism of xylophagous termites greatly depends on the gut microflora, which secrete enzymes that act synergistically with the host endogenous enzyme complexes and aid in the complete degradation of lignocellulose ingested by the termites. Till date, very few studies have been undertaken to illustrate the gut yeast flora of wood-feeding termites and their possible biotechnological applications. In this study, we uncovered high diversity of yeasts from the gut of three different species of wood-feeding termites, which expands the inventory of yeasts species being isolated from different termite species. Our study also highlights that the isolation of specific yeast species depends upon the host termite species. We have also observed that the yeasts belonging to Ascomycetes group were efficient ethanol producer and could not produce enzymes like xylanase, while on the other hand yeasts belonging to Basidiomycetes could produce a considerably high amount of xylanase but could not produce ethanol. We could get a few yeasts, which could produce significant amounts of xylanase; however, none of our strains could produce cellulase. Our aim was to obtain yeasts which could produce both of these enzymes so that such yeast strains can be co-cultured with efficient ethanol producers and could be used for consolidate bioprocessing (CBP) of lignocellulosic biomass to produce ethanol; however, we probably need to carry out an extensive sampling of yeasts from varied termite species to obtain such a yeast in future. Our investigation reveals that the gut of wood-feeding termites is a reservoir of unexplored yeasts, which may have immense use in biotechnological industries for sustainable production of xylanase and ethanol.
Supplemental Material
Download MS Word (12.1 KB)Supplemental Material
Download TIFF Image (5.8 MB)Supplemental Material
Download MS Word (21.5 KB)Supplemental Material
Download MS Excel (32 KB)Supplemental Material
Download MS Word (18.2 KB)Acknowledgements
The authors are thankful to the Director, MACS’ Agharkar Research Institute for providing the necessary facility to carry out the research work.
Disclosure statement
The authors declare that there are no conflicts of interest. All the experiments undertaken in this study comply with the current laws of the country where they were performed.
Additional information
Funding
References
- Kurtzman CP, Fell JW, Boekhout T. The yeasts: a taxonomic study. 5th ed. Amsterdam (Netherlands): Elsevier; 2011.
- Starmer WT, Lachance MA. Yeast ecology. In: Kurtzman CP, Fell JW, Boekhout Hiroshima, editors. The yeasts. 5th ed. Amsterdam (Netherlands): Elsevier; 2011. p. 65–83.
- Suh SO, McHugh JV, Pollock DD, et al. The beetle gut: a hyperdiverse source of novel yeasts. Mycol Res. 2005;109(Pt 3):261–265.
- Suh SO, Marshall CJ, McHugh JV, et al. Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol Ecol. 2003;12(11):3137–3145.
- Suh SO, Gibson CM, Blackwell M. Metschnikowia chrysoperlae sp. nov., Candida picachoensis sp. nov. and Candida pimensis sp. nov., isolated from the green lacewings Chrysoperla comanche and Chrysoperla carnea (Neuroptera: Chrysopidae). Int J Syst Evol Microbiol. 2004;54(Pt 5):1883–1890.
- Blackwell M. Made for each other: ascomycete yeasts and insects. Microbiol Spectr. 2017;5:945–962.
- Brune A. Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol. 2014;12(3):168–180.
- Schäfer A, Konrad R, Kuhnigk T, et al. Hemicellulose-degrading bacteria and yeasts from the termite gut. J Appl Bacteriol. 1996;80(5):471–478.
- Prillinger H, Messner R, König H, et al. Yeasts associated with termites: a phenotypic and genotypic characterization and use of coevolution for dating evolutionary radiations in asco- and basidiomycetes. Syst Appl Microbiol. 1996;19(2):265–283.
- Handel S, Wang T, Yurkov AM, et al. Sugiyamaella mastotermitis sp. nov. and Papiliotrema odontotermitis f.a., sp. nov. from the gut of the termites Mastotermes darwiniensis and Odontotermes obesus. Int J Syst Evol Microbiol. 2016;66(11):4600–4608.
- Ali SS, Wu J, Xie R, et al. Screening and characterizing of xylanolytic and xylose-fermenting yeasts isolated from the wood-feeding termite, Reticulitermes chinensis. PLoS One. 2017;12(7):e0181141.
- Kumar S, Singh SP, Mishra IM, et al. Recent advances in production of bioethanol from lignocellulosic biomass. Chem Eng Technol. 2009;32(4):517–526.
- Ohkuma M, Yuzawa H, Amornsak W, et al. Molecular phylogeny of Asian termites (Isoptera) of the families Termitidae and Rhinotermitidae based on mitochondrial COII sequences. Mol Phylogenet Evol. 2004;31(2):701–710.
- Aamir S, Sutar S, Singh SK, et al. A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol Quarant. 2015;5(2):74–81.
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Lone MA, Wani MR, Sheikh SA, et al. Evaluation of cellulase enzyme secreted by some common and stirring Rhizoshere fungi of Juglans regia L.by DNS Method. J Enzym Research. 2012;3:18–22.
- Lara CA, Santos RO, Cadete RM, et al. Identification and characterisation of xylanolytic yeasts isolated from decaying wood and sugarcane bagasse in Brazil. Antonie Van Leeuwenhoek. 2014;105(6):1107–1119.
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31(3):426–428.
- Yan J, Wei Z, Wang Q, et al. Bioethanol production from sodium hydroxide/hydrogen peroxide-pretreated water hyacinth via simultaneous saccharification and fermentation with a newly isolated thermotolerant Kluyveromyces marxianu strain. Bioresour Technol. 2015;193:103–109.
- Borzani W, Schmidell W, Lima UA, et al. Biotecnologia industrial: engenharia bioquímica. Vol. 2. São Paulo: Edgard Blücher; 2001.
- Sharma S, Nandal P, Arora A. Ethanol production from NaOH pretreated rice straw: a cost effective option to manage rice crop residue. Waste Biomass Valor. 2019;10(11):3427–3434.
- Shanbhag RR, Sundararaj R. Host range, pest status and distribution of wood destroying termites of India. J Trop Asian Entomol. 2012; 2:12–27.
- Adsul MG, Bastawde KB, Gokhale DV. Biochemical characterization of two xylanases from yeast Pseudozyma hubeiensis producing only xylooligosaccharides. Bioresour Technol. 2009;100(24):6488–6495.
- Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev. 2005;29(1):3–23.
- Stevens BJH, Payne J. Cellulase and xylanase production by yeasts of the genus Trichosporon. J Gen Microbiol. 1977;100(2):381–393.
- Saha BC. Hemicellulose bioconversion. J Ind Microbiol Biotechnol. 2003;30(5):279–291.
- Pandey AK, Kumar M, Kumari S, et al. Evaluation of divergent yeast genera for fermentation-associated stresses and identification of a robust sugarcane distillery waste isolate Saccharomyces cerevisiae NGY10 for lignocellulosic ethanol production in SHF and SSF. Biotechnol Biofuels. 2019;12(1):40.
