Abstract
Global temperatures are steadily increasing, leading to significant changes in microbial diversity and ecology. In the present study, we isolated high-temperature-growing fungi and fungi-like group (Oomycota) strains from freshwater environments of Korea and identified them based on cultural, morphological, and multilocus phylogenetic analyses. As a result, we introduce Saksenaea (Fungi) isolates as a new species, Saksenaea longicolla sp. nov. and record Phytophthora chlamydospora and P. lagoariana (Oomycota) new to Korea. In the growth experiments, they exhibited high-temperature tolerance, which can grow at 35–40 °C but become inactive at 4 °C and below. This study confirms the presence of high-temperature-tolerant fungi and oomycetes in Korea and suggests that the Korean climate conditions are changing in favor of these species. This indicates that climate warming is altering microbial distributions in freshwater environments.
1. Introduction
Global warming is altering microbial ecology, diversity [Citation1–3], physiology [Citation4–6], and interaction with abiotic and biotic factors [Citation4,Citation7,Citation8]. For terrestrial Fungi and Oomycota (previously classified under Fungi, but now under Straminipila), climate-changing effects have been established concerning their growth, reproduction, physiology, and distributions [Citation9–13]. However, the impact on the aquatic ecosystem, which is one of the important habitats for fungi and oomycetes, has been poorly studied, although it would result in changes in biotic communities and abiotic environmental factors [Citation14–16]. Fungi and oomycetes are abundant in aquatic ecosystems where they play key roles in food web dynamics and carbon cycle [Citation17–19]. They will give feedback to changes in climate gradient that may have a significant impact on aquatic environmental changes.
Ecological changes as a consequence of climate change on the Korean Peninsula are inevitable. According to the Intergovernmental Panel on Climate Change (IPCC), Korea can expect extended dry periods with warmer temperatures and frequent extreme precipitation events [Citation20–22]. As such, the subtropical zone in Korea is expanding, which will produce significant changes in the ecosystem. Interestingly, extreme cold events are also becoming more frequent [Citation23], caused by the loss of Arctic sea ice and the change in the Arctic atmosphere [Citation24–26]. This unprecedented phenomenon may give rise to considerable changes in Korea’s ecosystems. To date, there were relatively few studies focusing on these ecological changes and even fewer on fungal and oomycete communities despite their crucial ecological roles.
Saksenaea, a filamentous fungal genus characterized by large, flask-shaped sporangia, is affiliated with Mucorales [Citation27]. Since its first description from forest soil in India [Citation27], members of Saksenaea have been isolated mainly in soil habitats, but also diverse environments such as on the coast in Costa Rica [Citation28] and Taiwan [Citation29], a banana plantation in Honduras [Citation30], a groundnut field in Israel [Citation31], a pineapple field in Okinawa, Japan [Citation32], tree nurseries in Georgia, USA [Citation33], and forests in Panama [Citation34] and Brazil [Citation35]. S. dorisiae was recently described in freshwater [Citation36]. This species has been found only in the temperate continental region (the Republic of Serbia), while other members of Saksenaea described so far have been isolated in subtropical and tropical regions. Serbia is one of the countries experiencing a warming trend with climate change [Citation37], but it seems to be premature to mention any effect of climatic change on S. dorisiae because this species has been discovered in well water at a depth of 65 m [Citation36]. It should be noted that Saksenaea species are often involved in severe human and animal mucormycosis in tropical and subtropical regions [Citation38–40], certainly made possible because of their high-temperature tolerance, growing at 35–40 °C [Citation35,Citation41].
The genus Phytophthora belongs to the order Peronosporales of the Oomycota. Phytophthora is currently categorized in ten clades, according to their phylogenetic relationships, morphological and physiological characteristics [Citation42]. Many species of Phytophthora are well known, such as the pathogens which cause sudden oak death [Citation43], alder dieback [Citation44], root rot [Citation45], and late blight of potatoes and tomatoes [Citation46]. Phytophthora species occur abundantly also in riparian ecosystems, irrespective of disease [Citation47–51]. Especially, two phylogenetic clades, 6 and 9, of Phytophthora [Citation42,Citation52,Citation53] include many saprotrophic species that are abundantly distributed in littoral zones of rivers and lakes, or forested streams [Citation48,Citation49,Citation51,Citation54]. They play important roles on food webs in aquatic ecosystems as a decomposer of plant debris or prey for zooplankton by trophic transfer to higher levels [Citation19]. A unique part of these two clades is that they are characterized in terms of containing high-temperature-tolerant species [Citation19,Citation42,Citation48,Citation49,Citation55]. Their optimum temperature ranges mostly from 25 to 30 °C, but they are capable of growing at 35–40 °C. The high-temperature-tolerant species have been discovered mostly in hot climates, or where the littoral zones can reach high temperatures [Citation48,Citation55–57], although there are some wide temperature-tolerant species within clade 6, e.g., P. chlamydospora [Citation58], P. riparia [Citation59], which have often been recorded even in a cold climate.
Our microbial survey discovered high-temperature-growing fungal and oomycete isolates from freshwater, soil sediment, and decaying plant leaf matter originating from the reservoirs and mountain streams of Korea. These isolates were investigated for cultural and morphological features, and their optimum and minimal/maximal temperatures for growth were determined. Molecular sequencing analyses were carried out to determine their phylogenetic positions using the internal transcribed spacer (ITS) and large subunit (LSU) rDNA, and translation elongation factor 1 (TEF1α) regions for Saksenaea, and cytochrome c oxidase subunit I (cox1) and subunit II (cox2) mtDNA regions for Phytophthora. As a result, a novel species of Saksenaea and two species of Phytophthora, previously unrecorded in Korea, are reported in the present study.
2. Materials and methods
2.1. Isolation of fungal and oomycete strains
Fungal and oomycete isolates were collected from freshwater environments in Korea, including water, soil sediment, and decaying plant leaves. The collection details are outlined in , and climate details for the collection sites are given in . For Saksenaea isolates, a dilution plating method was used for soil samples at a 1:10 dilution. The diluted soil suspension was spread onto solid surfaces of two agar media: potato dextrose agar (PDA; Difco, Detroit, MI, USA) and V8 agar (V8A; 200 mL clarified V8 juice, 10 g CaCO3, 15 g agar, 800 mL deionized water). The growth of bacteria and other fungi was suppressed by adding antibiotics to the media of Saksenaea isolates 5 ppm benomyl, 25 ppm nystatin, 100 ppm penicillin G, 10 ppm pimaricin, 100 ppm streptomycin, and 100 ppm chloramphenicol. For Phytophthora isolates, water samples were distributed to both agar plates using a simple plating technique. The decaying plant leaves were cut into 3–5 mm2 pieces after washing with distilled water and were placed on both types of solid growth media. Bacterial and fungal growth was suppressed by adding 15 ppm rifampicin and 20 ppm nystatin to the media of Phytophthora isolates. The inoculated plates were incubated for 3 days at 25 °C in the dark. From the outgrowing mycelia, the new hyphal tips were isolated and transferred onto new agar plates. Representative cultures were deposited at the Nakdonggang National Institute of Biological Resources (NNIBR) or the Korean Agricultural Culture Collection (KACC).
Table 1. Collection details and GenBank accession numbers for Saksenaea and Phytophthora isolates investigated in the present study.
2.2. Morphological analysis
Cultural characteristics were investigated 3 days after inoculation of the isolates onto PDA, V8A, corn meal agar (CMA; Difco), malt extract agar (MEA; Difco), and Czapek solution agar (CZA; Difco). Microscopic structures were observed and photographed using an Olympus BX53F microscope (Olympus, Tokyo, Japan) equipped with a DigiRetina 16 M digital camera (Tucsen, Fuzhou, China), and a Leica M205C microscope (Leica, Wetzlar, Germany) equipped with a Dhyana 400DC camera (Tucsen).
2.3. Determination of temperature range for radial growth
To determine the optimal and minimal/maximal temperatures for growth, three strains (Sak-07, W655, and W694) were inoculated by transferring five replicates for each isolate onto four different media (PDA, V8A, MEA, and CZA) at 4, 10, 15, 20, 25, 30, 35, 40, and 45 °C, with additional temperatures of 37 and 42 °C for the Saksenaea strain (Sak-07). The colony radial growth was measured every day for 7 days and expressed as mm/day. Relative growth rates and standard deviation were calculated in Microsoft Excel.
2.4. DNA extraction, amplification, and sequencing
Genomic DNA of the strains was extracted using the MagListo 5 M plant Genomic DNA Extraction Kit (Bioneer, Daejeon, Korea), according to the manufacturer’s instructions. A MM400 mixer mill (Retsch GmbH, Haan, Germany) with glass beads was used to disrupt their culture tissues at 30 Hz/s for 15 min. PCR amplification of Saksenaea strains was performed for ITS region with primers ITS1 and ITS4 [Citation60], LSU rDNA region with NL-1 and NL-4 [Citation61], and TEF1α with MEF-11 and MEF-41 [Citation62]. For Phytophthora strains, two mitochondrial genes, cox1 and cox2 mtDNA, were amplified with two oomycete-specific primer sets, OomCox1-levup and OomCox1-levlo [Citation63] for cox1, and cox2-F [Citation64] and cox2-RC4 [Citation65] for cox2. DNA amplicons were sequenced by Macrogen Inc. (Seoul, Korea) after their purification using the AccuPrep PCR Purification Kit (Bioneer).
2.5. Phylogenetic analysis
All sequences were edited using the DNAStar software package 5.05 (DNAStar, Inc., Madison, WI, USA). A BLASTn search was carried out to investigate the sequence similarities with their reference sequences in NCBI GenBank. Multilocus phylogenetic analysis was performed using a combined dataset of TEF1α, ITS, and LSU rDNA for Saksenaea isolates and using a dataset of cox1 and cox2 mtDNA sequences for Phytophthora. In addition, the reference sequences of the type of authentic isolates of Saksenaea and Phytophthora were included in each dataset. The sequences of the individual marker were aligned using MAFFT 7 [Citation66] and then concatenated in SequenceMatrix v1.7.8 [Citation67]. Phylogenetic trees were constructed in MEGA 7.0 [Citation68]. For Saksenaea isolates, minimum evolution (ME) using the Kimura-2 model and maximum likelihood (ML) inferences using the General Time Reversible model were performed. For Phytophthora isolates, the Tamura-Nei model was used for both ME and ML analyses. Bootstrapping analysis was performed with 1000 replicates.
3. Results
3.1. Cultural and morphological analyses
Colonies of five Saksenaea strains (C17, Sak-06, Sak-07, Sak-19, and Sak-21) formed a radiate pattern on five agar media (PDA, V8A, MEA, CMA and CZA) at 25 °C in the dark and grew colorless with aerial hyphae (); aerial hyphae were denser on PDA, V8A, and MEA than on CMA and CZA. Radial growth with a circular layer was observed on the reverse side of the colony on PDA. Hyphae were hyaline, sparsely septate, and smooth. Asexual structures, including sporangiophores, sporangia, and sporangiospores, formed only on CZA. The sporangiophores were erect, unbranched, and formed singly. The sporangia were hyaline, flask-shaped, multi-spored, and developed terminally. The sporangiospores were mostly bacilliform or narrow middle rod-shaped, but rarely trapezoid. The cultural and morphological characteristics were close to those of S. dorisiae, S. oblongispora, and S. trapezispora. However, the sporangiophores of S. trapezispora were significantly longer than those of other species, including the Korean strains. The sporangia of S. oblongispora were smaller than those of S. dorisiae and the Korean ones. Saksenaea dorisiae was distinguished from the present strains by the narrow widths of sporangiophores, neck, and sporangia, and the somewhat smaller size of sporangiospores. The distinguishing morphological features between the Korean isolates and the closely related species of Saksenaea are summarized in .
Figure 1. Cultural and morphological characteristics of Saksenaea longicolla sp. nov. NNIBRFG21789 (SAK-07) on PDA (A, B), V8A (C, D), CMA (E, F), MEA (G, H), and CZA (I, J) after 72 h at 25 °C (A, C, E, G, I: observed view; B, D, F, H, J: reverse view). Microscopic structures: sporangiophore under a stereoscopic microscope (K, L) and under a light microscope (M, N), sporangiospores (O, P). Scale bar = 50 μm for K–N, 5 μm for O and P.
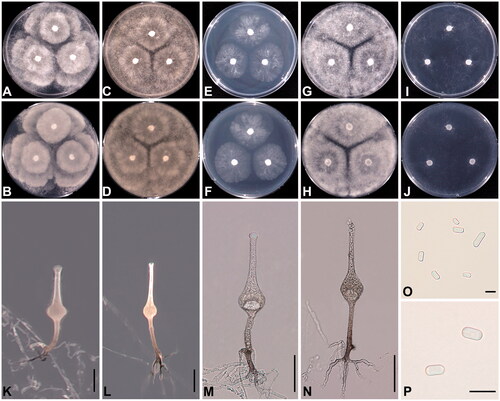
Table 2. Morphology and hyphal growth temperature of Saksenaea strain Sak-07 and their morphologically close species.
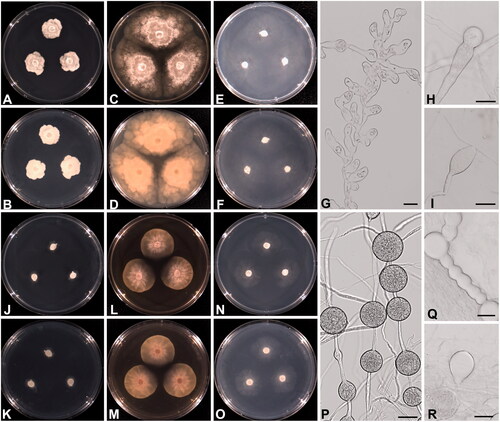
All Phytophthora isolates grew colorless on PDA, V8A, CMA, MEA, and CZA at 25 °C in the dark, with fewer aerial hyphae. Colonies of W694 formed a radiate pattern on all media. Sporangia were non-papillate, ovoid to obpyriform, formed on unbranched sporangiophores, and displayed commonly internal proliferation. Chlamydospores formed either terminally or intercalary and were observed abundantly at 25–30 °C. These characteristics were identical to those in the original description of Phytophthora chlamydospora [Citation69]. The strains of W655 and W675 were identified as Phytophthora lagoariana based on the colony growth pattern and morphological features described by Wallace [Citation70]. The colonies formed a vague rosaceous pattern on PDA, V8A, and MEA, but a radiate pattern on CMA and CZA. The strains grew submerged on CMA and CZA, while they formed a colorless mat on PDA, V8A, and MEA. These strains produced ovoid to obpyriform, non-papillate, and internally proliferating sporangia. Hyphal swelling was often observed, and chlamydospores were rarely produced. The colony patterns of P. chlamydospora (W694) and P. lagoariana (W655) on PDA, V8A, and CMA, along with morphological characteristics, are shown in .
Figure 2. Cultural and morphological characteristics of Phytophthora lagoariana NNIBRFG9322 (W655) (A–I), P. chlamydospora NNIBRFG9321 (W694) (J–R) on PDA (A, B, J, K), V8A (C, D, L, M), and CMA (E, F, N, O) after 72 h at 25 C° (A, C, E, J, L, N: observed view; B, D, F, K, M, O: reverse view). Microscopic structures observed under a light microscope: hyphal swellings (G, H), and sporangium (I) of P. lagoariana; chlamydospores (P, Q), and sporangium (R) of P. chlamydospora. Scale bar = 13.4 μm for G and H, 27 μm for I, P-R.
3.2. Phylogenetic analysis
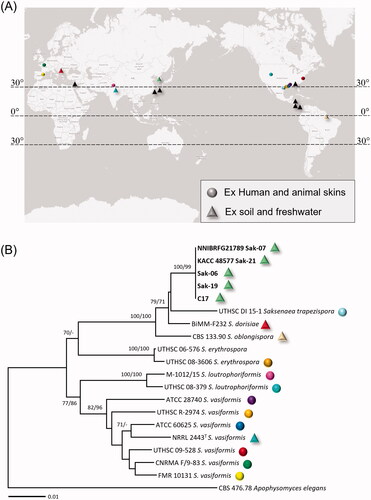
Based on a BLASTn search at the NCBI GenBank, the Korean isolates of Saksenaea were determined to be closest to S. trapezispora (UTHSC DI 15-1), with sequence similarities of 90% (229/254 bp) in ITS rDNA, 98% (687/703 bp) in LSU rDNA, and 99% (493/498 bp) in TEF1α. ME and ML analyses of a concatenated alignment of ITS, LSU, and TEF1α sequences were performed to infer the phylogenetic relationships between the Korean isolates and previously published Saksenaea species. As the ME and ML trees were congruent, only the ME tree is shown in . The multilocus tree revealed that the Korean isolates formed a distinct clade with high bootstrapping values in ME (100%) and ML (99%) analyses. They further grouped with S. dorisiae, S. oblongispora, and S. trapezispora, with the maximum supporting values in both analyses.
Figure 3. (A) Geographic distribution of Saksenaea species. Circle means the records of Saksenaea species or isolates from infected humans and animals. Triangle means the isolates from soil and freshwater, and black triangles mean the isolates with no sequence data in GenBank. (B) Multi-gene phylogenetic tree of Saksenaea species from the minimum evolution analysis of a concatenated alignment of three loci (TEF1α, ITS, LSU rDNA). Bootstrapping values (minimum evolution BP/maximum likelihood BP) higher than 70% were given above or below the branches (1,000 replicate). The strains isolated in Korea are shown in bold. Apophysomyces elegans was used as an outgroup. The scale bar equals the number of nucleotide substitutions per site.
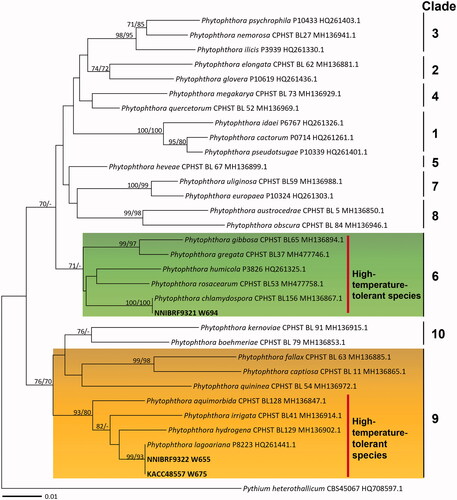
In BLASTn searching results for Phytophthora isolates, the cox2 (538 characters) and cox1 (640 characters) sequences of the Korean isolate W694 were identical to the authentic isolate CPHST BL156 (MH136867.1) of Phytophthora chlamydospora in cox1 sequence and P3176 (JF771548.1) in cox2. The isolates W655 and 675 matched the isolate P8223 (HQ261441.1 in cox1, HM534974.1 in cox2) of Phytophthora lagoariana with sequence similarities of 100% (634/634 bp) in cox1 and 99.8% (535/536 bp) in cox2. To infer the phylogenetic relationship between the authentic isolates of ten previously established clades of Phytophthora and the Korean isolates, ME and ML trees were constructed based on cox1 and cox2 mtDNA sequences. The phylogenetic tree based on the cox1 dataset is shown in . As the topologies constructed by both analyses were congruent, only the ME tree is presented. Three Korean isolates were placed in two different clades of Phytophthora, Clade 6 and Clade 9, with high supporting values. The strain W694 fell within Phytophthora Clade 6, which tolerates high temperatures, and further matched P. chlamydospora but was separated from non-chlamydospore-forming species [Citation48,Citation49]. The isolates of P. lagoariana, containing the Korean strains W655 and W675, formed a group with high-temperature-tolerant species of Clade 9. The optimum temperatures for these species range between 25 °C and 30 °C, but they can also grow at 35–40 °C [Citation42,Citation55].
Figure 4. Phylogenetic tree of Phytophthora species from the minimum evolution analysis based on cytochrome oxidase subunit I mtDNA sequences. Bootstrapping values (minimum evolution BP/maximum likelihood BP) higher than 70% were given above or below the branches (1,000 replicate). The strains isolated in Korea are shown in bold. Pythium heterothallicum was used as an outgroup. The scale bar equals the number of nucleotide substitutions per site.
3.3. Temperature range for radial growth
The strain Sak-07 of Saksenaea exhibited an optimum growth temperature ranging from 25 °C to 37 °C, with the lowest temperature at 10 °C (reaching 20–30 mm in diameter after a week) and the highest at 40 °C (). No growth was observed at 4 °C and 42 °C. Mean colony radial growth at 25 °C was observed to be 13.3 mm/day on PDA, V8A, and CZA, but 10 mm/day on MEA. On V8A and PDA, the colony grew as fast at 30–37 °C as at 25 °C. On MEA and CZA, the growth rate was lower at 37 °C than at 25–35 °C. The growth at 15 °C was moderately fast (reaching 40–50 mm in 4 days).
Figure 5. Relative growth rates of Saksenaea longicolla sp. nov. NNIBRFG21789 (SAK-07) for seven days at 4, 10, 15, 20, 25, 30, 35, 37, 40, 42, and 45 °C on four media, V8A (A), PDA (B), MEA (C), and CZA (D).
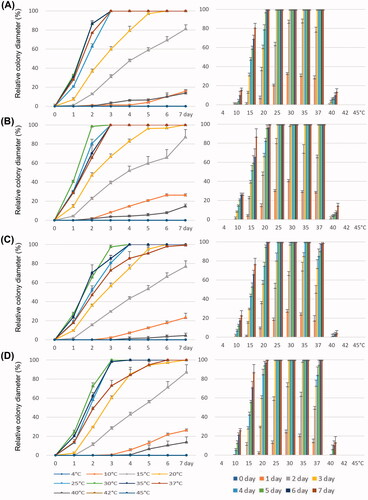
For P. lagoariana strain W655, the optimum temperature ranged from 25 °C to 35 °C (). At 25 °C, the strain grew fastest on V8A, with an average radial growth of 6.7 mm/day, but slowly on PDA (2.4 mm/day), MEA (0.9 mm/day), and CZA (1.1 mm/day). At 40 °C, the growth rate slowed down to 1.5 mm/day (V8A), 0.4 mm/day (PDA), and 0.2 mm/day (CZA), and no growth was observed on MEA. At 10 °C, it showed limited growth on V8A and PDA and no growth on MEA and CZA.
Figure 6. Relative growth rates of Phytophthora lagoariana W655 (A1–A4) and P. chlamydospora W694 (B1–B4) for seven days at 4, 10, 15, 20, 25, 30, 35, 40, and 45 °C on four media, V8A (A1, B1), PDA (A2, B2), MEA (A3, B3), and CZA (A4, B4).
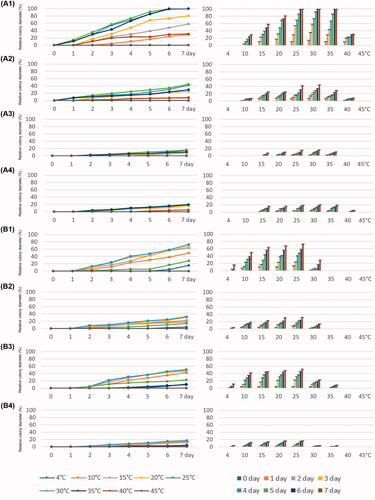
Compared to the W655, P. chlamydospora stain W694 grew substantially slower on all media (). The maximum growth temperature was 35 °C on MEA and CZA, and 30 °C on V8A and PDA. The optimum temperature for W694 ranged from 20 °C to 25 °C on all culture media. It was capable of growing at 4 °C (average 0.5 mm/day) as well as 10 °C (average 1.8 mm/day) on all culture media, although the growth rates were significantly lower. The growth rate of W694 was the highest at 25 °C on V8A. The colony radial growth on V8A was observed to be 4 mm/day at 25 °C. At 10 °C (2.6 mm/day) and 30 °C (1.4 mm/day); the colonies grew faster on V8A and MEA than on other media. At 30 °C on CZA, the strain showed limited growth (0.25 mm/day).
3.4. Taxonomy
Saksenaea longicolla D.J. Lee, B. Nam, & Y.J. Choi, sp. nov.
MycoBank: MB 838397
Etymology
From Latin “longus + collum”, characterizing the long neck of sporangiophores.
Description
Colonies reaching 65–80 mm after 4 days of incubation on CZA at 37 °C, whitish, with wispy aerial mycelium and abundant sporangiophores. Colonies on V8A, PDA, and MEA at 37 °C showing floccose and whitish, with no sporulation. Hyphae sparsely septate, hyaline, branched, thin-walled, and measuring 10–12 μm wide. Sporangiophores erect, generally arising singly, brown, unbranched, and measuring (58)78–135(168) (average 107) μm long and (5.1)6.6–13.9(21.7) (average 10) μm wide, with distinct spinulose stipe and a profuse dichotomously branched rhizoidal complex. Sporangia terminal, multispored, hyaline, flask-shaped, asperulate, and measuring (57)73–152(198) (average 113) μm long and (23)32–54(74) (average 43) μm wide. Neck measuring (35)53–114(151) (average 83) μm long, (6.8)7.3–12.7(8.1) (average 10) μm wide, with the apex closed with a mucilaginous plug but dissolved when mature. Sporangiospores are mostly bacilliform but rarely trapezoid, somewhat narrower at the middle and measuring (5.0)5.6–6.8(7.9) (average 6.2) μm long, (2.5)2.8–3.5(3.9) (average 3.1) μm wide. Zygospores were not observed.
Holotypus
Korea; Jeollabuk-do; Gunsan-si; Miryong-dong (35°57’01”N 126°40’47”E), ex soil sediment in a freshwater reservoir, Sept. 18, 2018, D.-J. Lee and Y.-J. Choi, NNIBRFG21789 (=Sak-07).
Notes
Like other members of Saksenaea, S. longicolla is high-temperature tolerant. In terms of being unable to grow above 42 °C, S. longicolla is close to its phylogenetically related species, S. trapezispora, S. oblongispora, and S. dorisiae [Citation35,Citation71]. However, the optimum (25–37 °C) and the maximum (40 °C) growth temperatures of S. longicolla are higher than those of S. trapezispora and S. oblongispora. In addition, S. longicolla grows slowly at 10 °C, while the latter two species are inactive [Citation35,Citation72]. Morphologically, Saksenaea longicolla is similar to S. trapezispora [Citation72] and S. oblongispora [Citation35], but distinguishable by the size of the sporangiophores, sporangia, and sporangiospores. Both BLASTn-based comparison and phylogenetic analysis of multilocus sequences supported this morphological identification. Saksenaea stains have been isolated mainly from the soil of diverse environments such as forest, beach, and grain and fruit fields, as well as animal skin infections. Similarly, S. longicolla was isolated from soil sediments of a freshwater reservoir. All species of Saksenaea described so far have a tropical and subtropical distribution in America [Citation28,Citation30,Citation33,Citation73,Citation74], Australia [Citation75,Citation76] and southern Asia [Citation29,Citation32,Citation38,Citation77,Citation78]. A warm-temperate site from which S. dorisiae has been recorded, the Republic of Serbia is experiencing warming trends [Citation37], similar to a Mediterranean climate. Additionally, as mentioned above, S. dorisiae has been discovered in the water at a depth well [Citation36], which is less affected by the air temperature. To date, there has been no record of Saksenaea species from cooler regions. Interestingly, the sporangiospores of S. dorisiae have been reported to be lively at low temperatures in water [Citation36], which could provide a hint that S. longicolla originating from freshwater could survive the cold season of Korea. However, further research is needed to answer this question.
Phytophthora chlamydospora
Brasier, C. and E. Hansen, North American Fungi 10 (2): 3 (2015) [MB#809175].
Description
Colonies growing colorlessly and slowly on PDA, V8A, and CMA at 25 °C, with few aerial mycelia, showing a radiate pattern on CMA and V8A, submerged growth on CMA, petaloid on V8A. Colony diameter after 72 h reaching 20–25 mm on PDA, 30–35 mm on V8A, and 25–30 mm on CMA. Sporangiophores hyalin, unbranched. Sporangia sometimes sympodial, obpyriform or ovoid, often elongated, non-papillate, and measuring 45–65 µm long, 30–40 µm wide; internal proliferation. Chlamydospores intercalary or terminal, occasionally in sessile form and measuring 16–35 µm in diameter.
Isolate examined
Korea; Jeollabuk-do; Imsil-gun; Seongsu-myeon; Seongsu-ri (35°38’04”N, 127°24’52”E), ex freshwater, 5 Sept 2018, B. Nam and Y.-J. Choi (NNIBRFG9321=W694).
Notes
The present study combining morphological and phylogenetic data revealed that the isolate W694 is identical to Phytophthora chlamydospora [Citation69]. The name P. chlamydospora was previously known informally as “P. taxon Pgchlamydo”, but Hansen et al. [Citation69] have redesigned this species based on a distinguishing feature of chlamydospore forming at a higher temperature. The Korean isolate also formed chlamydospores abundantly at 25–30 °C but not at 20 °C and below. In the present study, growth experiments verified that the Korean P. chlamydospora can grow at 35 °C but not at 40 °C, overlapping with a previous report that this isolate is usually able to survive at 36–37 °C [Citation69]. Members of Phytophthora Clade 6, including P. chlamydospora, are high-temperature tolerant and related to freshwater environments [Citation79,Citation80], which is in line with the Korean one obtained from the freshwater of a mountain stream. Phytophthora chlamydospora is often found in irrigation water, rivers, streams, and riparian areas of forest in Australia [Citation81], North America [Citation51], Europe [Citation82], Africa [Citation50], and Asia [Citation83]. Notably, this species is known as a pathogen and a saprophyte; in North America [Citation84] and Turkey [Citation85], it has been reported as a cause of root and crown rot of almonds. Remarkably, these potentially pathogenic species have inhabited the Korean peninsula.
Phytophthora lagoariana
Wallace, Sydney F. Diversity of Phytophthora Species in Costa Rica’s Tropical Forest. University of Maryland (2015).
Description
Colonies growing colorlessly on PDA, V8A, and CMA at 25 °C. On V8A forming a colorless mat with limited surface mycelia and a vague rosaceous pattern. On PDA growing slowly, with a rosaceous pattern. On CMA forming a radiate pattern with some aerial mycelium. Colony diameter after 72 h reaching >70 mm on V8A, 20–25 mm on PDA, and 55–60 mm on CMA. Sporangiophores hyaline extended. Sporangia obpyriform or ovoid, often elongated, non-papillate, 40–56 µm long, 25–40 µm wide; internal proliferation. Hyphal swelling is often observed. Chlamydospores are rarely produced and measuring 21–33 µm in diameter.
Isolates examined
Korea; Chungcheongnam-do; Yeongi-gun; Seo-myeon; Gobok-ri (36°36’29”N, 127°14’47”E), ex freshwater, 1 Jun 2018, B. Nam and Y.-J. Choi (NNIBRFG9322 = W655). Korea; Jeollabuk-do; Namweon-si; Inwol-myeon; Inwol-ri (35°27’08”N, 127°35’30”E), ex a decaying leaf in freshwater, 5 Sept 2018, B. Nam and Y.-J. Choi (KACC48557 = W675).
Notes
The isolates W655 and W675 were morphologically and phylogenetically identified as P. lagoariana [Citation70]. These isolates fell within Phytophthora Clade 9, of which many species are high-temperature tolerant but also well adapted to aquatic environments, e.g., P. hydrogena [Citation55], P. irrigata [Citation86], and P. aquimorbida [Citation56]. Similarly, the Korean isolates exhibited a high temperature tolerance, with an optimum growth temperature of 10–35 °C and were isolated from a freshwater reservoir and mountain stream. This species has been previously isolated from aquatic environments in the Cuyabeno Reserve in Ecuador and Carara National Park in Costa Rica [Citation70]. This is the first record of P. lagoariana in a nontropical region.
4. Discussion
Here, we report that high-temperature-tolerant fungus (Saksenaea) and oomycetes (Phytophthora) are present in the temperate but cold winter areas of Korea. Saksenaea species and Phytophthora lagoariana has previously been reported only in the tropics and subtropics. It is still unclear how they are able to overcome the cold winter of Korea. In the present growth experiment, S. longicolla did not grow at 4 °C and below, and P. lagoariana stopped growing even at 10 °C. However, the mean winter temperature of the sampling locations (Gunsan, Yeongi, Namwon, and Imsil) of the present study is below 4 °C, with the lowest temperature ranging from −10 to −20 °C and frequent frost events.
Their presence could be related to climate change as the warming in the Korean peninsula continues to accelerate. According to the Korea Meteorological Administration, Korea’s average temperature increased by 1.8 °C over the last century. Since 1910, Korea’s summer has increased by a month, from 80–110 to 110–140 days. In the sampling locations, the temperature has increased by around 1–2 °C for the past 50 years, and extreme heat events have become more frequent. This is an indication that the subtropical zone, restricted to the southern islands and coast of Korea, is expanding to the north.
Another crucial question is whether S. longicolla and two Phytophthora species are indigenous to Korea or exotic species. Given that they still prefer to grow at a much higher optimal temperature (25–35 °C) than the yearly mean temperatures (12.0–13.5 °C) of Korea, it seems likely that they may have recently immigrated from other (sub-)tropical or temperate regions, and then climate warming of Korea has made it easier for them to settle there. In addition, their ability to grow over a broad spectrum of temperatures (10–40 °C for S. longicolla and P. lagoariana and 4–35 °C for P. chlamydospora) may have facilitated more rapid adaptation.
It seems certain that the Korean peninsula’s climate conditions are changing in favor of high-temperature-tolerant fungus and oomycetes, which could encourage their presence and dominance in the ecosystems. The freshwater environment is an essential habitat for fungus and oomycetes, which are influential as saprophytes and parasites in aquatic food web structure and dynamics. Their diversity and distribution patterns are impacted by environmental settings with climatic factors, e.g., temperature, precipitation [Citation17,Citation87] that can be variable in global climate change. Understanding their diversity and distribution patterns under climate change is essential, and further research focusing on their ecological and functional traits is required.
Author contributions
Conceived and designed the experiments: B.N., D.J.L. and Y.J.C.; Performed the experiments: B.N. and D.J.L.; Analyzed and interpreted the data: B.N., D.J.L. and Y.J.C.; Wrote the article: B.N., D.J.L. and Y.J.C. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download MS Word (14.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Sequence data analyzed during the present study are available in GenBank.
Additional information
Funding
References
- Putten WHVd. Climate change, aboveground-belowground interactions, and species' range shifts. Annu Rev Ecol Evol Syst. 2012;43(1):365–383.
- Shade A, Peter H, Allison SD, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417.
- Singh BK, Bardgett RD, Smith P, et al. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8(11):779–790.
- Classen AT, Sundqvist MK, Henning JA, et al. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: what lies ahead? Ecosphere. 2015;6(8):1–12.
- Barcenas-Moreno G, Gomez-Brandon M, Rousk J, et al. Adaptation of soil microbial communities to temperature: comparison of fungi and bacteria in a laboratory experiment. Global Change Biol. 2009;15(12):2950–2957.
- Schimel J, Balser TC, Wallenstein M. Microbial stress-response physiology and its implications for ecosystem function. Ecology. 2007;88(6):1386–1394.
- Compant S, Sessitsch A, Van Der Heijden MGA. Climate change effects on beneficial plant–microorganism interactions. FEMS Microbiol Ecol. 2010;73(2):197–214.
- Larionova AA, Yevdokimov IV, Bykhovets SS. Temperature response of soil respiration is dependent on concentration of readily decomposable C. Biogeosciences. 2007;4(6):1073–1081.
- Andrew C, Halvorsen R, Heegaard E, et al. Continental‐scale macrofungal assemblage patterns correlate with climate, soil carbon and nitrogen deposition. J Biogeogr. 2018;45(8):1942–1953.
- Mohan JE, Cowden CC, Baas P, et al. Mycorrhizal fungi mediation of terrestrial ecosystem responses to global change: mini-review. Fungal Ecol. 2014;10:3–19.
- Boddy L, Büntgen U, Egli S, et al. Climate variation effects on fungal fruiting. Fungal Ecol. 2014;10:20–33.
- Scott J, Burgess T, Hardy G, et al. Climate modelling to determine the impacts of Phytophthora cinnamomi under future climate scenarios. Western Australia: Centre for Phytophthora Science and Management; 2013.
- Ghini R, Hamada E, Bettiol W. Climate change and plant diseases. Sci Agric. 2008;65:98–107.
- Boyero L, Pearson RG, Gessner MO, et al. A global experiment suggests climate warming will not accelerate litter decomposition in streams but might reduce carbon sequestration. Ecol Lett. 2011;14(3):289–294.
- Woodward G, Perkins Daniel M, Brown Lee E. Climate change and freshwater ecosystems: impacts across multiple levels of organization. Philos Trans R Soc Lond B Biol Sci. 2010;365(1549):2093–2106.
- Döll P, Zhang J. Impact of climate change on freshwater ecosystems: a global-scale analysis of ecologically relevant river flow alterations. Hydrol Earth Syst Sci. 2010;14(5):783–799.
- Grossart H-P, Van den Wyngaert S, Kagami M, et al. Fungi in aquatic ecosystems. Nat Rev Microbiol. 2019;17(6):339–354.
- Grossart H-P, Rojas-Jimenez K. Aquatic fungi: targeting the forgotten in microbial ecology. Curr Opin Microbiol. 2016;31:140–145.
- Marano A, Jesus A, De Souza J, et al. Ecological roles of saprotrophic Peronosporales (Oomycetes, Straminipila) in natural environments. Fungal Ecol. 2016;19:77–88.
- Song H-J, Sohn B-J. An evaluation of WRF microphysics schemes for simulating the warm-type heavy rain over the Korean peninsula. Asia-Pacific J Atmos Sci. 2018;54(2):225–236.
- Lee M-H, Ho C-H, Kim J, et al. Assessment of the changes in extreme vulnerability over East Asia due to global warming. Clim Change. 2012;113(2):301–321.
- Jung I-W, Bae D-H, Kim G. Recent trends of mean and extreme precipitation in Korea. Int J Climatol. 2011;31(3):359–370.
- Kim Y, Lee S. Trends of extreme cold events in the Central regions of Korea and their influence on the heating energy demand. Weather Clim Extremes. 2019;24:100199.
- Kim B-M, Son S-W, Min S-K, et al. Weakening of the stratospheric polar vortex by arctic sea-ice loss. Nat Commun. 2014;5(1):4646.
- Francis JA, Vavrus SJ. Evidence linking arctic amplification to extreme weather in mid-latitudes. Geophys Res Lett. 2012;39(6):L06801.
- Honda M, Inoue J, Yamane S. Influence of low arctic sea-ice minima on anomalously cold Eurasian winters. Geophys Res Lett. 2009;36(8):L08707.
- Saksena SB. A new genus of the Mucorales. Mycologia. 1953;45(3):426–436.
- Claudette L. M., Caballero M., I. S, editors. Microorganism infection of olive ridley eggs. the 12th Annual Workshop on Sea Turtle Biology and Conservation; 1992; Jekyll Island, Georgia: NOAA Technical Memorandum NMFSSEFSC.28.
- Cheen C-Y. Education in medical mycology of Taiwan. Nihon Ishinkin Gakkai Zasshi. 1987;28(1):32–38.
- Goos RD. Further observations on soil fungi in Honduras. Mycologia. 1963;55(2):142–150.
- Joffe AZ, Borut SY. Soil and kernel mycoflora of groundnut fields in Israel. Mycologia. 1966;58(4):629–640.
- Watanabe T. Pictorial atlas of soil and seed fungi: morphologies of cultured fungi and key to species. 3 ed. Watanabe T, editor. Boca Raton, Florida: CRC press; 2010.
- Hodges CS. Fungi isolated from Southern Forest tree nursery soils. Mycologia. 1962;54(3):221–229.
- Farrow WM. Tropical soil fungi. Mycologia. 1954;46(5):632–646.
- Alvarez E, Garcia-Hermoso D, Sutton DA, et al. Molecular phylogeny and proposal of two new species of the emerging pathogenic fungus saksenaea. J Clin Microbiol. 2010;48(12):4410–4416.
- Labuda R, Bernreiter A, Hochenauer D, et al. Saksenaea dorisiae sp. nov., a new opportunistic pathogenic fungus from Europe. Int J Microbiol. 2019;2019:6253829.
- Vukovic A, Vujadinovic M, Rendulic S, et al. Global warming impact on climate change in Serbia for the period 1961–2100. Therm Sci. 2018;22(6 Part A):2168–2267.
- Baradkar VP, Mathur M, Taklikar S, et al. Fatal rhino-orbito-cerebral infection caused by Saksenaea vasiformis in an immunocompetent individual: first case report from India. Indian J Med Microbiol. 2008;26(4):385–387.
- Gkegkes ID, Kotrogiannis I, Konstantara F, et al. Cutaneous mucormycosis by Saksenaea vasiformis: an unusual case report and review of literature. Mycopathologia. 2019;184(1):159–167.
- Relloso S, Romano V, Landaburu MF, et al. Saksenaea erythrospora infection following a serious sailing accident. J Med Microbiol. 2014;63(2):317–321.
- Kaufman L, Padhye AA, Parker S. Rhinocerebral zygomycosis caused by Saksenaea vasiformis. J Med Vet Mycol. 1988;26(4):237–241.
- Martin FN, Abad Z, Balci Y, et al. Identification and detection of Phytophthora: reviewing our progress, identifying our needs. Plant Dis. 2012;96(8):1080–1103.
- Davidson JM, Werres S, Garbelotto M, et al. Sudden oak death and associated diseases caused by Phytophthora ramorum. Plant Health Prog. 2003;4(1):12.
- Brasier CM, Kirk SA, Delcan J, et al. Phytophthora alni sp. nov. and its variants: designation of emerging heteroploid hybrid pathogens spreading on alnus trees. Mycol Res. 2004;108(10):1172–1184.
- Erwin DC, Ribeiro OK. Phytophthora diseases worldwide. St. Paul (MN): American Phytopathological Society (APS Press); 1996.
- Nowicki M, Foolad MR, Nowakowska M, et al. Potato and tomato late blight caused by Phytophthora infestans: an overview of pathology and resistance breeding. Plant Dis. 2012;96(1):4–17.
- Hansen EM, Reeser PW, Sutton W. Phytophthora beyond agriculture. Annu Rev Phytopathol. 2012;50:359–378.
- Jung T, Stukely MJC, Hardy GESJ, et al. Multiple new Phytophthora species from ITS clade 6 associated with natural ecosystems in Australia: evolutionary and ecological implications. Persoonia. 2011;26:13–39.
- Nagel J, Gryzenhout M, Slippers B, et al. Characterization of Phytophthora hybrids from ITS clade 6 associated with riparian ecosystems in South Africa and Australia. Fungal Biol. 2013;117(5):329–347.
- Oh E, Gryzenhout M, Wingfield BD, et al. Surveys of soil and water reveal a goldmine of Phytophthora diversity in South African natural ecosystems. IMA Fungus. 2013;4(1):123–131.
- Reeser PW, Sutton W, Hansen EM, et al. Phytophthora species in Forest streams in Oregon and Alaska. Mycologia. 2011;103(1):22–35.
- Blair JE, Coffey MD, Park SY, et al. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol. 2008;45(3):266–277.
- Kroon LP, Brouwer H, de Cock AW, et al. The genus Phytophthora anno 2012. Phytopathology. 2012;102(4):348–364.
- Nechwatal J, Mendgen K. Widespread detection of Phytophthora taxon salixsoil in the Littoral zone of Lake Constance, Germany. Eur J Plant Pathol. 2006;114(3):261–264.
- Yang X, Gallegly ME, Hong C. A high-temperature tolerant species in clade 9 of the genus Phytophthora: P. hydrogena sp. nov. Mycologia. 2014;106(1):57–65.
- Hong C, Richardson PA, Hao W, et al. Phytophthora aquimorbida sp. nov. and Phytophthora taxon 'aquatilis' recovered from irrigation reservoirs and a stream in Virginia, USA. Mycologia. 2012;104(5):1097–1108.
- Hüberli D, Hardy GSJ, White D, et al. Fishing for Phytophthora from Western Australia’s waterways: a distribution and diversity survey. Australasian Plant Pathol. 2013;42(3):251–260.
- Sims LL, Sutton W, Reeser P, et al. The Phytophthora species assemblage and diversity in riparian alder ecosystems of Western Oregon, USA. Mycologia. 2015;107(5):889–902.
- Hansen EM, Reeser PW, Sutton W. Phytophthora borealis and Phytophthora riparia, new species in Phytophthora ITS clade 6. Mycologia. 2012;104(5):1133–1142.
- White T, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA GD, Sninsky JJ, White TJ, editor. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–322.
- Kurtzman CP, Robnett CJ. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5' end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35(5):1216–1223.
- O'Donnell K, Lutzoni F, J. Ward T, et al. Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia. 2001;93(2):286–296.
- Bala K, Robideau G, Désaulniers N, et al. Taxonomy, DNA barcoding and phylogeny of three new species of Pythium from Canada. Persoonia. 2010;25:22–31.
- Hudspeth DSS, Nadler SA, Hudspeth MES. cox2 molecular phylogeny of the Peronosporomycetes. Mycobiology. 2000;92(4):674.
- Choi Y-J, Beakes G, Glockling S, et al. Towards a universal barcode of oomycetes-a comparison of the cox1 and cox2 loci. Mol Ecol Resour. 2015;15(6):1275–1288.
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780.
- Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27(2):171–180.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874.
- Hansen E, Reeser P, Sutton W, et al. Redesignation of Phytophthora taxon pgchlamydo as Phytophthora chlamydospora sp. nov. N Am Fungi. 2015;10(2):1–14.
- Wallace SF. Diversity of Phytophthora species in Costa Rica's Tropical Forest. [Master Thesis]: University of Maryland 2015.
- Crous PW, Wingfield MJ, Burgess TI, et al. Fungal planet description sheets: 558–624. Persoonia. 2017;38:240–384.
- Crous PW, Wingfield MJ, Burgess TI, et al. Fungal planet description sheets: 469–557. Persoonia. 2016;37:218–403.
- Blanchet D, Dannaoui E, Fior A, et al. Saksenaea vasiformis infection, French Guiana. Emerg Infect Dis. 2008;14(2):342–344.
- Vega W, Orellana M, Zaror L, et al. Saksenaea vasiformis infections: case report and literature review. Mycopathologia. 2006;162(4):289–294.
- Stewardson AJ, Holmes NE, Ellis DH, et al. Cutaneous zygomycosis caused by Saksenaea vasiformis following water-related wound in a 24-year-old immunocompetent woman. Mycoses. 2009;52(6):547–549.
- Wilson PA. Zygomycosis due to Saksenaea vasiformis caused by a magpie peck. Med J Aust. 2008;189(9):521–522.
- Padhye AA, Koshi G, Anandi V, et al. First case of subcutaneous zygomycosis caused by Saksenaea vasiformis in India. Diagn Microbiol Infect Dis. 1988;9(2):69–77.
- Tanphaichitr VS, Chaiprasert A, Suvatte V, et al. Subcutaneous mucormycosis caused by Saksenaea vasiformis in a thalassaemic child: first case report in Thailand. Mycoses. 1990;33(6):303–309.
- Brasier CM, Cooke DE, Duncan JM, et al. Multiple new phenotypic taxa from trees and riparian ecosystems in Phytophthora gonapodyides-P. megasperma ITS clade 6, which tend to be high-temperature tolerant and either inbreeding or sterile. Mycol Res. 2003;107(3):277–290.
- Brasier CM, Hamm PB, Hansen EM. Cultural characters, protein patterns and unusual mating behaviour of Phytophthora gonapodyides isolates from Britain and North America. Mycol Res. 1993;97(11):1287–1298.
- Burgess TI, Webster JL, Ciampini JA, et al. Re-evaluation of Phytophthora species isolated during 30 years of vegetation health surveys in Western Australia using molecular techniques. Plant Dis. 2009;93(3):215–223.
- Jung T, Blaschke M. Phytophthora root and collar rot of alders in Bavaria: distribution, modes of spread and possible management strategies. Plant Pathol. 2004;53(2):197–208.
- Huai W-X, Tian G, Hansen EM, et al. Identification of Phytophthora species baited and isolated from Forest soil and streams in northwestern Yunnan province, China. For Path. 2013;43(2):87–103.
- Browne GT, Ott NJ, Forbes H, et al. First report of Phytophthora chlamydospora causing crown and root rot on almond in California. Plant Dis. 2020;104(7):2033–2033.
- Türkölmez Ş, Derviş S, Çiftçi O, et al. First report of Phytophthora chlamydospora causing root and crown rot on almond (Prunus dulcis) trees in Turkey. Plant Dis. 2016;100(8):1796–1796.
- Hong C, Gallegly ME, Richardson PA, et al. Phytophthora irrigata, a new species isolated from irrigation reservoirs and Rivers in Eastern United States of America. FEMS Microbiol Lett. 2008;285(2):203–211.
- Redondo MA, Boberg J, Stenlid J, et al. Contrasting distribution patterns between aquatic and terrestrial Phytophthora species along a climatic gradient are linked to functional traits. Isme J. 2018;12(12):2967–2980.
