Abstract
The species within the family Cunninghamellaceae during an investigation of soil microfungi in Korea, in which three strains were isolated from Gangwon, Chungbuk, and Gyeongbuk provinces, designated as KNUF-22-121A, KNUF-22-126A, and KNUF-22-316, respectively. Because the morphological and molecular analyses of these three strains were identical, KNUF-22-316 underwent further detailed study. Phylogenetic analyses based on the concatenated nucleotide sequences of the internal transcribed spacer region and the large subunit 28S rRNA gene revealed that the strain belonged to the genus Absidia, but occupied a distinct phylogenetic position. The strain KNUF-22-316 was compared with closely related species Absidia radiata CGMCC 3.16257T and Absidia yunnanensis CGMCC 3.16259T, morphologically different with shorter sporangiophores, smaller sporangia and columellae, and the consistent presence of collars. Here, we provide a detailed description and images of this proposed new species, which we have named Absidia microsporangia sp. nov.
1. Introduction
The number of circumscribed genera in the family Cunninghamellaceae (Mucorales, Mucoromycetes, Mucoromycota) is controversial. Initially, it included three genera: Cunninghamella, Sigmoideomyces, and Thamnocephalis [Citation1]. Currently, six genera are reported in the family Cunninghamellaceae including Absidia, Chlamydoabsidia, Cunninghamella, Gongronella, Halteromyces, and Hesseltinella [Citation2]. Absidia, the type of the family Cunninghamellaceae, was first reported by Van Tieghem [Citation3]. To date, genus Absidia contains 51 species and members of the genus are characterized by (i) sporangiophores arranged singly or in pairs or groups on stolons, (ii) rhizoids at both ends of stolons and never opposite the sporangiophores, (iii) deliquescent-walled and apophysate sporangia, (iv) columellae bearing one to several projections, and (v) zygospores enclosed by appendages [Citation2,Citation4,Citation5]. The genus Absidia includes various fungal species that are found throughout the environment, most of which are ubiquitous in soil, dung, insects, leaf litter, food, air, and particularly in the mycangia of ambrosia beetles as well as the body surface of bats [Citation6]. Furthermore, species within Absidia possess important metabolites for industrial and medical applications, such as steroids, α-galactosidase, laccase, fatty acids, and chitosan [Citation7].
This study aimed to expand our knowledge of native fungal species diversity in Korea and the use of fungal resources in various industries. Here, we present the morphological and molecular characteristics of strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316, belonging to the genus Absidia.
2. Materials and methods
2.1. Sample collection and fungal strain isolation
The fungal strains used in this investigation were isolated from soil samples collected from Gangwon (37°07′32.4″N, 129°09′16.3″E), Chungbuk (36°42′37.6″N, 127°39′05.5″E), and Gyeongbuk (35°18′35.83″N, 129°12′10.75″E) provinces, Korea. Isolation was performed using the plate dilution method as previously described [Citation8]. Strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 showed similar cultural characteristics and were from different provinces. Thus, they were selected for further morphological and molecular phylogenetic analyses.
2.2. Cultural and morphological characterization
Strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 were cultured on potato dextrose agar (PDA; Difco, Detroit, MI) and malt extract agar (MEA; Difco, Detroit, MI) for seven days at 25 °C to investigate their cultural and morphological characteristics [Citation9,Citation10]. Cultural characteristics such as color, shape, and size, were observed. Morphological characteristics were examined under light microscopy (BX-50, Olympus, Tokyo, Japan).
2.3. Genomic DNA extraction, polymerase chain reaction (PCR) amplification, and sequencing
Total genomic DNA was extracted from fungal mycelia of strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 cultured on PDA plates using a HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, South Korea) according to the manufacturer’s instructions. The internal transcribed spacer (ITS) regions and partial sequences of the large subunit (LSU) 28S rRNA gene were amplified using the primer pair NS5M and LR5M [Citation11]. Then, the PCR products were purified using ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA) and sequenced by SolGent (Daejeon, South Korea) using primers ITS1F, ITS4, and LR0R [Citation12–14].
2.4. Molecular phylogenetic analyses
Strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 underwent phylogenetic analysis along with sequences retrieved from the National Center for Biotechnology Information GenBank database (). Ambiguous regions were deleted from alignments, and evolutionary distance matrices for the neighbor-joining algorithm were calculated using the Kimura two-parameter model [Citation15,Citation16]. Phylogenetic relationships were inferred by tree topology using maximum likelihood [Citation17] and maximum parsimony methods with MEGA7.0 software and 1000 bootstrap replications [Citation18,Citation19].
Table 1. GenBank accession numbers of sequences used for the phylogenetic analyses in this study.
Table 2. Morphological comparison between Absidia microsporangia sp. nov. (KNUF-22-316AT) and the phylogenetically closest species of Absidia.
3. Results
3.1. Taxonomy
Strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 were found to be morphologically identical, and they clustered together with respect to molecular phylogeny. Thus, these three strains were described as a new species. Since they were identical, only the cultural and morphological characteristics of strain KNUF-22-316 were described in this study.
Absidia microsporangia S.K. Lim, S.Y. Lee and H.Y. Jung sp. nov. ()
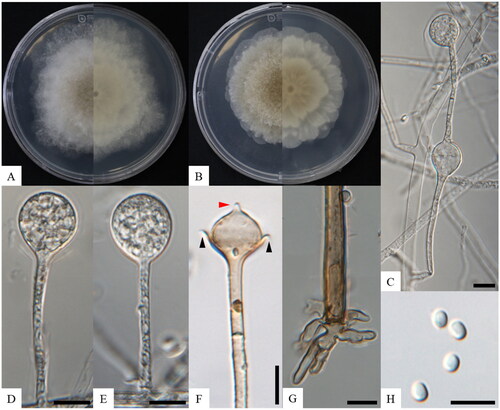
Figure 1. Morphology of Absidia microsporangia sp. nov. KNU-22-316T. (A, B) Colonies on PDA and MEA at 25 °C for seven days, obverse and reverse, respectively; (C) swelling on sporangiophores; (D, E) sporangium; (F) columellae, collars, and projections; (G) rhizoid; (H) sporangiophores. Scale bars: C–I = 10 μm. Black arrows, collars; a red arrow, projection.
MycoBank: 853470
Etymology: The specific epithet is derived from Latin “microsporangia”, referring to the small size of sporangia.
Typus: The strain was isolated from soil containing plant debris collected in May 2022 from Korea, Gyeongbuk province, Gijang-gun, Daleum mountain (35°18′35.83″N, 129°12′10.75″E). The stock culture of type strain KNUF-22-316T (NIBRFGC000509838) was deposited in the National Institute of Biological Resources as a metabolically inactive culture.
Habitat and distribution: Rhizosphere soil containing plant debris collected from Gangwon (Gagok-myeon, Samcheok-si), Chungbuk (Jeungpyeong-eup, Jeungpyeong-gun), and Gyeongbuk (Jeonggwan-eup, Gijang-gun) provinces, Korea.
Cultural characteristics: Colonies on PDA at 25 °C for seven days moderately fast growth, attaining 73.0–83.2 mm in diameter, floccose, initially white, soon becoming gray to brown, irregular edges; reverse white, irregular zonate (). Colonies on MEA at 25 °C for seven days moderately fast growth, attaining 53.6–58.6 mm in diameter, floccose, initially white, soon becoming brown, irregular edges; reverse white, irregular zonate ().
Morphological characteristics: Hyphae are branched, hyaline at first, brownish when mature, aseptate when juvenile, septate with age, and 2.4–12.4 µm (n = 30) in diameter. Sporangiophores arise from stolons, mostly erect, bent, in whorls of 1–3, branched, monopodial or sympodial, hyaline, and 36.1–102.8 µm × 1.8–3.1 µm (n = 30). Swellings are usually present below the sporangia, oval to pyriform, hyaline, often with a septum (). Sporangia are oval to subglobose, hyaline when young, dark green when old, smooth, multi-spored, and 9.2–16.6 µm × 8.4–16.6 µm (n = 30) (). Apophyses are distinct, hyaline or subhyaline, gradually widening upwards (base: 1.8–2.6 µm, top: 3.7–5.0 µm, n = 30). Collars are present. Columellae are subglobose or oval, hyaline, subhyaline, or light brown, and 6.2–11.8 µm × 5.6–11.7 µm (n = 30). Projections are single, hyaline to light brown, and 1.2–2.6 µm long (n = 30) (). Sporangiospores are regular in size and shape, mostly oval or subglobose, hyaline, smooth-walled, and 2.9–5.8 µm × 2.0–4.4 µm wide (x̅ = 4.0–3.0, n = 70) (). Rhizoids are root-like, often unbranched, and well-developed (). Chlamydospores are absent. Zygospores are not observed.
Other materials examined: Korea, Samcheok-si, Gagok-myeon, Deokpung valley (37°07′32.4″N, 129°09′16.3″E), on soil containing plant debris, May 2021, KNUF-22-121A; Korea, Jeungpyeong-gun, Jeungpyeong-eup, Jwagu mountain (36°42′37.6″N, 127°39′05.5″E), on soil containing plant debris, May 2021, KNUF-22-126A.
Note: The colonies of the strain KNUF-22-316 grew to 59 mm at 25 °C for seven days, whereas both the phylogenetically closest strains, Absidia radiata CGMCC 3.16257T and A. ampullacea CGMCC 3.16054T, reached 65 mm in the same conditions. Morphologically, the strain KNUF-22-316 produced shorter sporangiophores (36.1–102.8 µm × 1.8–3.1 µm) than A. radiata CGMCC 3.16257T (45.0–273.0 × 3.0–5.0 µm), A. ampullacea CGMCC 3.16054T (30.0–320.0 × 2.5–5.5 µm), and Absidia yunnanensis CGMCC 3.16259T (29.0–159.0 × 2.0–5.0 µm) [Citation2,Citation10]. Additionally, sporangia produced by KNUF-22-316 (9.2–16.6 × 11.5–16.6 µm) were smaller than those of A. radiata CGMCC 3.16257T (17.5–33.5 × 18.5–30.0 µm), A. ampullacea CGMCC 3.16054T (17.5–37.5 × 17.5–45.0 µm), and A. yunnanensis CGMCC 3.16259T (16.0–27.5 × 16.5–27.0 µm) [Citation2,Citation10]. Collars were absent in A. radiata CGMCC 3.16257T and A. ampullacea CGMCC 3.16054T, sometimes present in A. yunnanensis CGMCC 3.16259T, and consistently present in strain KNUF-22-316 [Citation2,Citation10]. The columellae of strain KNUF-22-316 (6.2–11.8 µm × 5.6–11.7) were smaller compared with those of A. radiata CGMCC 3.16257T (13.5–22.5 × 14.0–24.0 µm), A. ampullacea CGMCC 3.16054T (8.5–22.5 × 10.5–24.0 µm), and A. yunnanensis CGMCC 3.16259T (6.5–18.5 × 7.5–22.0 µm) [Citation2,Citation10]. Thus, strain KNUF-22-316 (representative of the proposed new species) differed from the closest species (A. radiata CGMCC 3.16257T, A. ampullacea CGMCC 3.16054T, and A. yunnanensis CGMCC 3.16259T) in terms of sporangiophores, sporangia, columellae, and the presence or absence of collars ().
3.2. Phylogenetic analysis
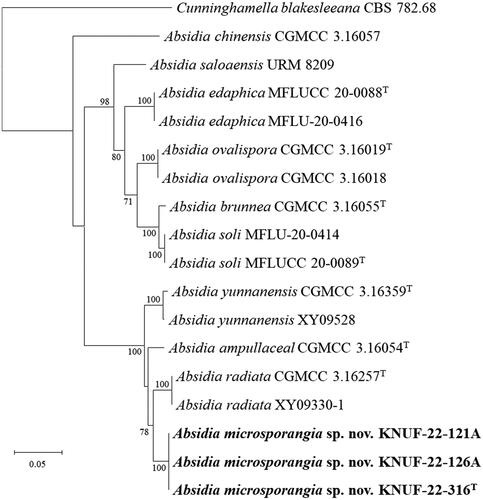
The sequence lengths of the ITS regions and LSU gene for all strains of KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 were 515 and 907 bp, respectively. The ITS regions of KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 showed a sequence similarity of 94.5%, 89.9%, and 88.6% with A. radiata CGMCC 3.16257T, A. yunnanensis CGMCC 3.16259T, and A. ampullacea CGMCC 3.16054T, respectively. For the LSU gene sequences, the three strains revealed a high similarity of 98.8% with A. ampullacea CGMCC 3.16054T, 97.7% similarity with A. yunnanensis CGMCC 3.16259T, and a 97.5% similarity with A. radiata CGMCC 3.16257T. A multilocus sequence analysis was performed using concatenated ITS regions and partial LSU gene sequences of strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 (). The neighbor-joining phylogenetic tree based on the concatenated sequences revealed that strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 occupied a distinct cluster from other Absidia species (). Based on phylogenetic analysis and morphological observations, strains KNUF-22-121A, KNUF-22-126A, and KNUF-22-316 were distinct from the previously identified Absidia species. Thus, they should be considered a novel species in the genus Absidia, for which we have proposed the name Absidia microsporangia sp. nov.
Figure 2. Neighbor-joining phylogenetic tree based on ITS regions and LSU gene sequences showing the phylogenetic position of the three isolated strains among the related strains in the genus Absidia. Cunninghamella elegans CBS 167.53 was used as the outgroup. The strains isolated in this study are indicated in bold, and the numbers above the branches represent the bootstrap values (>70%) obtained for 1000 replicates. Scale bar = 0.02 substitutions per nucleotide position.
4. Discussion
Since 2015, 27 out of a total of 51 species of Absidia have been reported, outnumbering those described in the last century. The species of Absidia are well known as ubiquitous in soil, dung, and decaying plants, as well as insect remains. However, most Absidia species including recently reported new strains of the genus Absidia such as A. abundans, A. lobata, A. radiata, A. sichuanensis, and A. yunnanensis, were isolated from soil [Citation10]. Additionally, A. jindoensis was isolated from rhizosphere soil of pine trees in Korea [Citation20]. Although, an increasing number of Absidia species, their ecological distribution has still not been unraveled [Citation7]. Despite obscure ecological distribution, the genus Absidia was well known for its industrial application by secondary metabolites [Citation21]. For example, A. spinosa has been reported to decolorize effluents by degrading cresol red, a toxic dye used in the textile industry [Citation22], while A. cylindrospora was found to remove trace metals, such as Cu, Cd, and Pb, present in soil [Citation23] and produce proteases, which are enzymes of biotechnological interest with high commercial value, using coffee residue as a substrate [Citation24]. Thus, the report of A. microsporangia sp. nov. isolated from soil could contribute to enlarging our knowledge of the ecological distribution of the genus Absidia and have potential industrial applications by its secondary metabolites.
In conclusion, this research presents our discovery of a new species, A. microsporangia sp. nov., isolated from a soil sample in Korea and provides a description according to phylogenetic and morphological evidence. The findings highlight the diversity within the genus Absidia and its ecological significance. Additionally, the potential industrial applications of Absidia species, particularly through secondary metabolites, underscore the importance of this fungus. Therefore, further research is needed to reveal the ecological distribution of the genus Absidia and the industrial potential of secondary metabolites produced by Absidia microsporangia sp. nov.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Benjamin RK. The merosporangiferous Mucorales. Aliso. 1959;4(2):321–433. doi: 10.5642/aliso.19590402.05.
- Zhao H, Nie Y, Zong TK, et al. Species diversity, updated classification and divergence times of the phylum Mucoromycota. Fungal Divers. 2023;123:49–157.
- Van Tieghem P. Troisiéme mémoire sur les Mucorinée. Ann Sci Nat Bot Ser 6. 1878;4:312–399.
- Hoffmann K. Identification of the genus Absidia (Mucorales, Zygomycetes): a comprehensive taxonomic revision. In: Gherbawy Y, Voigt K, editors. Molecular identification of fungi. Berlin/Heidelberg, Germany: Springer; 2010. p. 439–460.
- Zong TK, Zhao H, Liu XL, et al. Taxonomy and phylogeny of four new species in Absidia (Cunninghamellaceae, Mucorales) from China. Front Microbiol. 2021;12:677836. doi: 10.3389/fmicb.2021.677836.
- Cordeiro TRL, Nguyen TTT, Lima DX, et al. Two new species of the industrially relevant genus Absidia (Mucorales) from soil of the Brazilian Atlantic Forest. Acta Bot Bras. 2020;34:549–558.
- Zhao H, Nie Y, Zong TK, et al. Three new species of Absidia (Mucoromycota) from China based on phylogeny, morphology and physiology. Diversity. 2022;14(2):132. doi: 10.3390/d14020132.
- Das K, Ryu JJ, Hong SM, et al. Molecular phylogeny and morphology of Tolypocladium globosum sp. nov. isolated from soil in Korea. Mycobiology. 2023;51(2):79–86. doi: 10.1080/12298093.2023.2192614.
- Urquhart AS, Idnurm A. Absidia healeyae: a new species of Absidia (Mucorales) isolated from Victoria, Australia. Mycoscience. 2021;62(5):331–335. doi: 10.47371/mycosci.2021.06.001.
- Zhao H, Nie Y, Zong TK, et al. Species diversity and ecological habitat of Absidia (Cunninghamellaceae, Mucorales) with emphasis on five new species from forest and grassland soil in China. J Fungi. 2022;8:1–19.
- Zhao H, Lv ML, Liu Z, et al. High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. Bioenergy Res. 2021;14:1196–1206.
- White TJ, Bruns T, Lee S, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. PCR protocols: a guide to methods and applications. New York: Academic Press, Inc.; 1990. p. 315–322.
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118.
- Rehner SA, Samuels GJ. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol Res. 1994;98:625–634.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425.
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581.
- Posada D, Crandall KA. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;89(3):134–145. doi: 10.1007/BF01734359.
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–416.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Wanasinghe DN, Phukhamsakda C, Hyde KD, et al. Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89(1):1–236. doi: 10.1007/s13225-018-0395-7.
- Heidary M, Habibi Z. Microbial transformation of androst-4-ene-3, 17-dione by three fungal species Absidia griseolla var. igachii, Circinella muscae and Trichoderma virens. J Mol Catal B Enzym. 2016;126:32–36.
- Kristanti RA, Zubir MMFA, Hadibarata T. Biotransformation studies of cresol red by Absidia spinosa M15. J Environ Manag. 2016;172:107–111.
- Albert Q, Leleyter L, Lemoine M, et al. Comparison of tolerance and biosorption of three trace metals (Cd, Cu, Pb) by the soil fungus Absidia cylindrospora. Chemosphere. 2018;196:386–392. doi: 10.1016/j.chemosphere.2017.12.156.
- Cardoso KBB, Souza BKPA, Brandão-Costa RMP, et al. Waste coffee as an excellent substrate for collagenase production by filamentous fungi Cunninghamella phaeospora and Absidia cylindrospora. Act Sci Nutr Health. 2017;1:3–6.
